Abstract
Elastin-like polypeptides (ELPs) are biocompatible designer polypeptides with inverse temperature transition behavior in solution. They have a wide variety of possible applications and a potential medical importance. Currently, production of ELPs is done at lab scale in Escherichia coli shake flask cultures. With a view to future large scale production, we demonstrate secreted production of ELPs in methanol-induced fed-batch cultures of Pichia pastoris and purification directly from the culture medium. The production of ELPs by P. pastoris proved to be pH dependent within the experimental pH range of pH 3 to 7, as an increasing yield was found in cultures grown at higher pH. Because ELP produced at pH 7 was partly degraded, a pH optimum for production of ELP was found at pH 6 with a yield of 255 mg of purified intact ELP per liter of cell-free medium.
Similar content being viewed by others
Introduction
Elastin is present in elastic fibers within the extracellular matrix of vertebrates. It is produced and secreted from the cells as tropoelastin, a soluble protein consisting mainly of alternating hydrophilic and hydrophobic regions (Gacko 2000; Mithieux and Weiss 2005). Once secreted, tropoelastin is cross-linked at lysine residues within the hydrophilic regions to form an insoluble protein network with elastic properties originating from entropic recoil of extended hydrophobic regions (Bedellhogan et al. 1993; Mithieux and Weiss 2005). When a solution of tropoelastin is heated above a certain temperature, the hydrophobic regions cause contraction, desolvation, and aggregation of the protein. This reversible process, called inverse temperature transition, eventually results in phase separation of the solution (Cox et al. 1974; Urry 1976; Urry et al. 1974).
The inverse temperature transition has inspired scientists to create elastin-like polypeptides (ELPs) based on the amino acid repeat sequence found in hydrophobic regions. Several of these repeat sequences gave elastin-like thermoresponsive behavior in solution. The pentapeptide repeat sequence “VPGXG” (with any amino acid except proline at the X position) has become the most commonly used. The big advantage of ELPs over natural elastin sequences is a high degree of control over the transition temperature, which is dependent on the hydrophobicity of the amino acid at the X position and the length of the ELP. Placement of a more hydrophobic amino acid at the X position in VPGXG, or an increased length, results in a lower transition temperature (Urry et al. 1991a, b, 1992). An increased length of the ELP also reduces the concentration dependency of its transition temperature (Meyer and Chilkoti 2004). It is also possible to introduce a pH-dependent response by placement of charged amino acids at the X position because the transition temperature of charged and non-charged VPGXG repeats is dramatically different (Urry 1992).
This tunable transition temperature in combination with their biocompatible nature makes ELPs especially interesting for medical applications (Chilkoti et al. 2006; Rincon et al. 2006; Rodriguez-Cabello et al. 2007; Urry et al. 1991a, b). Some examples are drug targeting and gene delivery by use of local hyperthermia (Chen et al. 2008; Chilkoti et al. 2002), controlled drug delivery via bioresorption of ELP gels (Megeed et al. 2002), and tissue engineering with cross-linked ELP biomaterial as an implantable scaffold (McHale et al. 2005). A nonmedical application is the use of ELP as a fusion-tag for easy purification of target proteins by inverse transition cycling (ITC). ITC is a scalable and efficient purification method even at very low concentrations (Christensen et al. 2007; Ge and Filipe 2006; Ge et al. 2005; Gillies et al. 2008; Meyer and Chilkoti 1999; Trabbic-Carlson et al. 2004).
Currently, the most often used method for lab-scale production of ELPs is Escherichia coli shake flask cultures in combination with a hyper-expression protocol that gives typical yields of approximately 300 mg l−1 ELP (Guda et al. 1995; Trabbic-Carlson et al. 2003). Other expression hosts that have been used for expression of ELPs or natural elastin sequences are tobacco cell cultures (Zhang et al. 1995), cultures of Aspergillus nidulans (Herzog et al. 1997), and transgenic tobacco plants (Guda et al. 2000) and Saccharomyces cerevisiae (Toonkool and Weiss 2001). The common feature in these systems is that ELP is produced and recovered from inside the cells. We have studied secreted expression of ELPs by Pichia pastoris in fed-batch cultures because this species is well known for its high protein secretion capacity, and scale up of P. pastoris fed-batch cultures is relatively easy (Cereghino et al. 2002; Cregg et al. 2000). Furthermore, we successfully used this host for secreted expression of other repetitive polypeptides with a biased amino acid composition (Werten et al. 1999, 2001, 2008). In this work, we show that P. pastoris can be used for the secreted production of ELPs. To our knowledge, this is the first report of secreted ELP production by any expression host.
Materials and methods
Construction of strains
To create vector pMTL23-AIII, synthetic oligonucleotides AIII-FW and AIII-RV (Eurogentec, Table 1) were annealed to form a double-stranded adapter, which was ligated into EcoRI/XhoI-digested pMTL23 (Chambers et al. 1988) from which the normally present BsaI site had been removed. The same procedure was used to construct vector pMTL23-EX using oligonucleotides EX-FW and EX-RV (Eurogentec, Table 1).
The DNA fragment ELP10 was designed to encode 10 VPGXG pentapeptide repeats, with at the X position five valines, three alanines, and two glycines in quasi-random order. It was constructed by annealing ELPa and ELPb oligonucleotides into two dsDNA fragments (Eurogentec, Table 1) and jointly ligating these dsDNA fragments into BsmBI/EcoRI-digested oligomerization vector pMTL23-AIII so as to obtain pMTL23-AIII-ELP10.
A 90-mer of the VPGXG pentapeptide was subsequently constructed by recursive directional ligation (Meyer and Chilkoti 2002). The ELP10 fragment was cut from pMTL23-AIII-ELP10 with BsmFI/FokI and then ligated into a dephosphorylated BsmFI-linearized pMTL23-AIII-ELP10 vector, yielding pMTL23-AIII-ELP20. This process was repeated until pMTL23-AIII-ELP90 was obtained. The ELP90 fragment was then cut out of pMTL23-AIII-ELP90 with BsmFI/EcoRI and ligated into likewise digested pMTL23-EX. Finally, the ELP90 fragment was cloned into pPIC9 (Invitrogen) via EcoRI/XhoI, resulting in pPIC9-ELP90. This generates a fusion of ELP90 to an α-mating factor secretion signal that is proteolytically removed upon secretion. Genomic integration of SalI-linearized pPIC9-ELP90 into P. pastoris GS115 was achieved by electroporation as described before (Werten et al. 1999). When the final construct was sequenced, it was found that one glycine was missing in comparison with the designed sequence (Fig. 1). This difference is probably a result of synthesis errors in one of the oligonucleotides used, but cannot have an effect on the transition temperature of the ELP.
Protein expression
P. pastoris fed-batch fermentations for production of ELP were performed in 2.5 l Bioflo 3000 fermentors using BioCommand software for data acquisition and supervisory control (New Brunswick Scientific). The fermentation procedure was similar to a procedure with fixed methanol concentration described before (Zhang et al. 2000). The batch phase of the fermentation was started by addition of 125 ml preculture grown in minimal glycerol media to 1.25 l basal salt medium (BSM; Invitrogen) supplied with 5.5 ml PTM1 trace salts (Invitrogen) and adjusted to the final pH. When glycerol had run out, a glycerol fed batch was run with a constant glycerol feed of 20 g l−1 h−1 for 1 h. After the glycerol fed-batch phase, a transition phase was started by addition of methanol to a final concentration of 0.2%m/v and a programmed linear reduction of the glycerol feed to 0 g l−1 h−1 over 3 h. For the remainder of the fermentation, the methanol concentration was maintained at 0.2%m/v by an off-gas methanol sensor-controller. Biomass concentration of the cultures was determined daily by determining wet cell weight from 40 ml samples centrifuged at 3,500 rpm for 10 min in a Beckman Allegra-6 centrifuge with GH3.8A rotor.
Protein purification
After 48 h of induction, the fermentations were stopped, and cells were spun down at 4°C and 4560 RPM in a RC5C centrifuge (Sorvall) for 10 min using an SLA3000 rotor (Sorvall). The obtained cell-free medium was used for ELP purification by ITC. In the first ITC cycle, ELP was precipitated by an addition of NaCl to a concentration of 2 M and 30 min of incubation at 65°C followed by centrifugation at 45°C and 10,000 rpm for 60 min using an SLA3000 rotor. The ELP pellets were dissolved in a tenfold smaller volume of ice-cold Milli-Q water and centrifuged at 4°C for 45 min and 15,000 rpm in an SS34 rotor. The supernatant was then subjected to a second ITC cycle, in which both centrifugation steps (hot and cold) were carried out at 15,000 RPM for 45 min in an SS34 rotor. The obtained ELP solution was extensively dialyzed using Spectra/Por 7® tubing with a 1,000 Da molecular weight cutoff and subsequently freeze-dried for protein quantification by weighing.
Protein analysis
SDS-PAGE analysis of ELP expression was performed using a NuPAGE Novex system (Invitrogen) with 10% Bis–Tris gels and MES-SDS running buffer (Invitrogen) with Seeblue plus2 (Invitrogen) as a marker. The ELP bands were visualized by copper staining (Lee et al. 1987).
For N-terminal protein sequencing, the proteins were blotted onto a PVDF membrane using a Criterion blotter (Biorad) and CAPS transfer buffer (2.2 g CAPS in 10% methanol, pH 11). After staining, the blots with Coomassie Brilliant Blue ELP protein bands, appearing white on a blue background, were used for N-terminal sequencing by Midwest Analytical (St. Louis, MO, USA).
The amino acid composition of purified ELP was determined by amino acid analysis of freeze-dried protein samples by Ansynth Service B.V. (Roosendaal, the Netherlands). The analysis included detection of 17 common amino acids with a detection limit of 10 µmol g−1 (excluding tryptophan, cysteine, and methionine).
MALDI-TOF MS analysis of purified ELP was performed using an Ultraflex mass spectrometer (Bruker), with a 600 μm AnchorChip target (Bruker) and protein calibration standard II (Bruker) using 2,5-dihydroxyacetophenone as a matrix (Wenzel et al. 2006).
Proteolysis experiments
Extracellular proteolytic degradation of ELP was tested by incubating purified ELP with samples of cell-free medium and cell-containing broth. Intact ELP (produced at pH 6) was added to a final concentration of 400 mg l−1 to each of the samples and to Milli-Q water as a control. After overnight incubation at 30°C, ELP was recovered by a single ITC cycle and checked by SDS-PAGE gel for the presence of degradation fragments. Finally, the recovery of ELP was quantified by weighing of the product after freeze drying.
Transition temperature analysis
The transition temperature of ELP, defined as the point where turbidity reaches 50% of its maximum value, was determined by measuring absorbance with a UV1650PC spectrophotometer at 380 nm with a CPS240a temperature controller and UV-probe software (Shimadzu, ‘s-Hertogenbosch Netherlands). The measurements were conducted by heating 25 µM ELP in phosphate-buffered saline (NaCl 8 g l−1, KCl 0.2 g l−1, Na2HPO4 1.44 g l−1, KH2PO4 0.24 g l−1, pH 7.4) from 50°C to 58°C at 0.2°C per minute.
Results
Production and purification of ELP
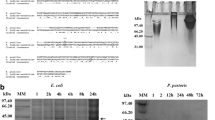
To determine the suitability of P. pastoris for the expression of ELP, an expression strain was generated by genomic integration of an ELP gene encoding 90 VPGXG repeats. More specifically, an ELP90 was selected in which the X position of the repeat consists for 50% of valine, 30% of alanine, and 20% of glycine placed in a quasi-random order. This ELP90 conformation was selected for its relevant transition temperature and the availability of literature data on both expression level in E. coli and transition temperature at various concentrations. Appropriate culture conditions for expression of ELP were determined in a series of fed-batch cultures with pH values ranging from 3 to 7. We found that even though at the high end (pH 5 to 7) precipitation of salts was apparent, there was no noticeable influence on culture growth, as all cultures grew to a similar cell density of 410–450 g l−1 wet cell weight 72 h after inoculation. At that time point, the fermentations were stopped, and ELP was purified from the broth by ITC and dialysis. It was found that the amount of ELP obtained per liter of supernatant increased from 0 mg l−1 at pH 3 to 410 mg l−1 at pH 7 (Fig. 2). SDS-PAGE analysis of the purified ELP showed that ELP produced at pH 4 to pH 6 yielded a single high intensity band migrating at 44 kDa, which corresponds to ELP of 35.4 kDa because ELP is known for a deviating migration speed (Meyer and Chilkoti 2002). The additional low intensity bands in the higher mass range are presumably caused by SDS-resistant ELP multimers. In addition, ELP produced at pH 7 also contained a ladder pattern of smaller bands (Fig. 3).
Product characterization
The amino acid content of ELP produced at pH 6 and pH 7 was determined, which confirmed that only alanine, proline, glycine, and valine were present at detectable levels in each sample. Moreover, in both samples, these four different amino acids were present in the approximate theoretical molar percentages found in ELP (Fig. 4). Linear least-squares fitting of the observed composition of each sample, with a combination of both the theoretical composition of ELP and an experimentally determined average composition of P. pastoris cell-free broth, indicated that >99% of the amino acids were derived from ELP. This suggests that the ladder pattern of bands found at pH 7 contains smaller ELP fragments (not other proteins) and that the low intensity bands of higher mass found at pH 4–6 also consist of ELP.
Mass spectrometry was performed with samples from ELP produced at pH 4, 5, and 6. The obtained spectra contain a main peak at 35.37 ± 0.01 kDa (Fig. 5), while the theoretical mass of ELP90 is 35.77 kDa. In agreement with SDS-PAGE, these spectra do not contain peaks at lower mass values. However, there were two smaller peaks visible next to the main peak, at approximately 35.49 and 35.69 kDa. An explanation for that was found in results of N-terminal sequencing of purified ELP produced at pH 6. Here, it was found that approximately 20% of all N-termini had the expected GPVPGGGV sequence, while approximately 80% of the proteins had an N-terminal sequence GGGVPGVG. Together with the MALDI results, this suggests that 80% of the produced ELP is missing four amino acids at the N-terminus (GPVP). Such an ELP would have a theoretical mass of 35.41 kDa, which is in reasonable agreement with the observed 35.34-kDa peak. These truncated N-termini probably resulted from unintended processing of -X-P- dipeptides by dipeptidyl amino peptidase (DPAPase). Although this activity has not been described for P. pastoris, it is known for other yeast species that DPAPase acts toward both -X-A-X-A- and -X-P-X-P- (Egel-Mitani and Hansen 1987; Kreil 1990; Matoba et al. 1988).
MALDI spectra of purified ELP produced at pH 7 contained multiple peaks in the lower mass range (Fig. 5). A consistent pattern of repeating peak triplets could be discerned at an interval of approximately 2 kDa. These regular intervals suggest that the ELP was cleaved by a protease at specific sites approximately 25 amino acids (or 5 VPGXG repeats) apart. In agreement, the N-terminal sequencing of both full size band and a pool of smaller bands of this ELP revealed that only four different N-termini were present: the unique N-terminal sequence GPVPGGGV that is only found in the true N-terminus of ELP and three other sequences (GGGVPGVG, GVPGVGVP, and GVPGGGVP). This limited number of N-termini and the pattern in MALDI spectra both indicated protease digestion at specific sites. Unfortunately, the repetitive amino acid sequence prevents from pinpointing the exact location cleavage within ELP.
The transition temperature of ELP produced at pH 6 was determined by turbidity measurements, and in good agreement with previously reported values of 50 –54°C, a transition temperature of 53.2°C was found (Fig. 6; Meyer and Chilkoti 2004).
Analysis of extracellular ELP stability and ITC performance at low pH
Two possible causes for the lower ELP yields at low pH were investigated: (1) complete degradation of part of the secreted ELP in cultures grown at pH 3–5 and (2) reduced efficiency of ITC at low pH. For the test, purified ELP (produced at pH 6) was added to a final concentration of 400 mg l−1 to both cell-free broth and cell-containing medium that were obtained from a fermentation at pH 3 with a strain devoid of recombinant ELP genes and to MQ water as a blank comparison. These samples were then incubated overnight at 30°C to mimic fermentation conditions, and afterward, ELP was purified from the samples by ITC. On SDS-PAGE, the intensity of full-size ELP bands in both samples was not drastically different from the blank comparison, and there were no signs of degradation (Fig. 7). Moreover, the amount ELP quantified after freeze drying was approximately 80% to 100% of the amount originally added, confirming that no important extracellular degradation of the product occurred at pH 3.
Discussion
Elastin-like polypeptides have been subject of research for approximately 45 years (Urry 1976; Urry et al. 1974). Within the last 20 years, the momentum of research has increased because the application of heterologous protein expression for production introduced a precise control over amino acid composition and polypeptide size (Cappello 1990; Heslot 1998). The heterologous expression of ELPs has been reported for several expression hosts, including bacteria (Guda et al. 1995; Trabbic-Carlson et al. 2003), fungi (Herzog et al. 1997; Toonkool and Weiss 2001), and plant cells or plants (Guda et al. 2000; Zhang et al. 1995). These studies have in common that the produced ELP remains contained within the cells. Here, we used P. pastoris for secreted expression of ELPs.
Because the pH is known to influence the proteolytic activity in high cell density fed-batch cultures of P. pastoris (Cregg et al. 2000; Jahic et al. 2003), the secreted expression of ELP was first tested within a pH range of pH 3 to 5. This is the pH range commonly used for P. pastoris fed-batch cultures, with pH boundaries set by reduced growth of P. pastoris below pH 3 and salt precipitation of basal salts media above pH 5 (Files et al. 2001; Zhang et al. 2007). When a yield was found that increased with pH, we decided to broaden the pH range to pH 3–7. In agreement with previous reports (Inan et al. 1999; Jahic et al. 2003; Sinha et al. 2005), this resulted in visible salt precipitation at both pH 6 and 7, but did not cause operational problems or nutrient starvation. There was a further increase in the amount of ELP at both pH 6 and pH 7, although at pH 7 the listed amount includes also smaller fragments. The experimental optimum for ELP production by P. pastoris is therefore pH 6.
Looking in more detail at the influence of pH on ELP production, we observed two distinct effects: (1) a gradual change of ELP yields over the full pH range and (2) a sudden appearance of smaller fragments at pH 7. Later on, we will discuss the gradual change of ELP yield, but first, the appearance of smaller fragments at pH 7 will be addressed. In general, proteolytic activity leads to pH-dependent degradation of secreted proteins; this degradation is usually observed by a reduced activity or yield of the produced enzyme or protein (Bollok et al. 2005; Clare et al. 1991; Inan et al. 1999). In our case, the ELP fragments still posses the characteristic temperature response, and therefore, they are extracted from the broth with the ITC purification method. After purification, the fragments are distinguishable on SDS-PAGE gels as a ladder pattern with distinct bands. Also, in MALDI-TOF spectra, the distinct fragment sizes are visible, in a repeating pattern of triplet peaks approximately 2 kDa apart. N-terminal sequencing detects only four different N-termini within the fragment pool. Because of the repetitivity of the amino acid sequence, it is impossible to state with certainty at which location cleavage takes place.
A second influence of pH is found in the gradual increase of ELP yield with rising pH. One reason why the gradual increasing yield was unexpected is that the pH is not documented to influence P. pastoris growth or AOX promoter-dependent protein expression (apart from pH-induced changes in extracellular protease degradation). That the expression level, obtained with the AOX promoter and the Pichia pastoris translation/transcription machinery, is not pH dependent is indicated by the fact that the yield of expression products, obtained with this system, is in some cases optimal at low pH, and in other cases optimal at high pH (Li et al. 2007; Mattanovich et al. 2004). We therefore assume the transcription/translation level of ELP to be largely independent of the extracellular pH. On the other hand, a pH-induced change in extracellular protease activity also seems unlikely because of the observed yield pattern. As described above, pH-induced changes in extracellular protease activity are commonly found in high cell density cultures of P. pastoris, resulting in degradation of the produced protein (Jahic et al. 2003; Sinha et al. 2005). In contrast, the ELP yield increases gradually from 0 to 255 mg l−1 in a pH ranging from 3 to 6, while there are no traces of degradation present. That would require complete degradation of only part of the secreted ELP while leaving the other part completely intact. Such a degradation pattern is highly improbable. Additional evidence is obtained by the experiment, performed to detect ELP-directed protease activity at pH 3. If extracellular protease degradation caused the yield gradient, we would expect the highest protease activity at pH 3. But, in the stability experiment, there was no degradation detected, and therefore, we conclude that extracellular protease activity is not causing the difference in yield. A reduced efficiency of ITC at low pH was another potential explanation that we tested. We found that the recovery of ELP from culture medium at pH 3 was 80–100% in the ELP stability experiment, showing that pH did not influence the recovery of ELP by ITC.
Because the pH did not cause degradation or reduced the recovery of secreted ELP and because the transcription/translation of the AOX promoter-controlled ELP genes is unlikely to be of influence, the secretory pathway of P. pastoris (including the periplasmic space) remains as the location where ELP yields were influenced by the (extracellular) pH. With this in mind, we hypothesize that the changes in the (extracellular) pH caused changes in the intracellular protease activity, the availability of chaperones, or the efficiency of ELP transport across the cell wall. This could result in aggregation, mislocalization, or intracellular degradation of only part of the expressed ELP. Additionally, this can also explain the seeming absence of similar pH dependencies in other P. pastoris expression studies, because they are potentially specific for ELP or a select group of proteins.
To conclude, we have shown here that ELP production by P. pastoris is possible, with yields similar to those obtained in E. coli. Unlike previous studies on ELP production using other hosts, the P. pastoris expression system described here secretes the ELP. This secretion simplifies the purification by eliminating the need for cell disruption. On a more general note, fed-batch fermentations of P. pastoris are also known for good scalability, which means that the observed yields (although subject to improvement) are indicative of results that can be obtained in future large-scale production (Zhao et al. 2008). We believe that the present work creates new possibilities for future expression studies of fusion proteins that combine ELP with a second functional group. Such combinations are necessary for most of the potential applications of ELP (Chow et al. 2008; Rodriguez-Cabello et al. 2007; Simnick et al. 2007). For some fusion proteins, secretory expression by P. pastoris merely adds an optional expression host, with a potential for high product yields. In other cases, secreted expression of ELP fusion proteins by P. pastoris can become advantageous, for example when disulfide bridges are present or when glycosylation is essential (Demain and Vaishnav 2009).
References
Bedellhogan D, Trackman P, Abrams W, Rosenbloom J, Kagan H (1993) Oxidation, cross-linking, and insolubilization of recombinant tropoelastin by purified lysyl oxidase. J Biol Chem 268:10345–10350
Bollok M, Henriksson H, Kallas A, Jahic M, Teeri TT, Enfors SO (2005) Production of poplar xyloglucan endotransglycosylase using the methylotrophic yeast Pichia pastoris. Appl Biochem Biotechnol 126:61–77
Cappello J (1990) The biological production of protein polymers and their use. Trends Biotechnol 8:309–311
Cereghino GPL, Cereghino JL, Ilgen C, Cregg JM (2002) Production of recombinant proteins in fermenter cultures of the yeast Pichia pastoris. Curr Opin Biotechnol 13:329–332. doi:https://doi.org/10.1016/S0958166902003300
Chambers SP, Prior SE, Barstow DA, Minton NP (1988) The pMTL nic - cloning vectors. I. Improved pUC polylinker regions to facilitate the use of sonicated DNA for nucleotide sequencing. Gene 68:139–149
Chen T-H, Bae Y, Furgeson D (2008) Intelligent biosynthetic nanobiomaterials (IBNs) for hyperthermic gene delivery. Pharmaceutical Res 25:683–691
Chilkoti A, Dreher MR, Meyer DE (2002) Design of thermally responsive, recombinant polypeptide carriers for targeted drug delivery. Adv Drug Deliv Rev 54:1093–1111
Chilkoti A, Christensen T, MacKay JA (2006) Stimulus responsive elastin biopolymers: Applications in medicine and biotechnology. Curr Opin Chem Biol 10:652–657. doi:https://doi.org/10.1016/j.cbpa.2006.10.010
Chow D, Nunalee ML, Lim DW, Simnick AJ, Chilkoti A (2008) Peptide-based biopolymers in biomedicine and biotechnology. Mater Sci Eng, R Rep 62:125–155
Christensen T, Trabbic-Carlson K, Liu W, Chilkoti A (2007) Purification of recombinant proteins from Escherichia coli at low expression levels by inverse transition cycling. Anal Biochem 360:166–168
Clare JJ, Romanos MA, Rayment FB, Rowedder JE, Smith MA, Payne MM, Sreekrishna K, Henwood CA (1991) Production of mouse epidermal growth-factor in yeast: high-level secretion using Pichia pastoris strains containing multiple gene copies. Gene 105:205–212
Cox BA, Starcher BC, Urry DW (1974) Coacervation of tropoelastin results in fiber formation. J Biol Chem 249:997–998
Cregg J, Cereghino J, Shi J, Higgins D (2000) Recombinant protein expression in Pichia pastoris. Mol Biotechnol 16:23–52
Demain AL, Vaishnav P (2009) Production of recombinant proteins by microbes and higher organisms. Biotechnol Adv 27:297–306
Egel-Mitani M, Hansen MT (1987) Nucleotide-sequence of the gene encoding the Saccharomyces kluyveri alpha-mating pheromone. Nucleic Acids Res 15:6303–6303
Files D, Ogawa M, Scaman CH, Baldwin SA (2001) A Pichia pastoris fermentation process for producing high-levels of recombinant human cystatin-C. Enzyme Microb Technol 29:335–340
Gacko M (2000) Elastin: Structure, properties and metabolism. Cell Mol Biol Lett 5:327–348
Ge X, Filipe CDM (2006) Simultaneous phase transition of ELP tagged molecules and free elp: an efficient and reversible capture system. Biomacromolecules 7:2475–2478. doi:https://doi.org/10.1021/bm060507n
Ge X, Yang DSC, Trabbic-Carlson K, Kim B, Chilkoti A, Filipe CDM (2005) Self-cleavable stimulus responsive tags for protein purification without chromatography. J Am Chem Soc 127:11228–11229
Gillies A, Hsii J, Oak S, Wood D (2008) Rapid cloning and purification of proteins: gateway vectors for protein purification by self-cleaving tags. Biotechnol Bioeng 101:229–240
Guda C, Zhang X, McPherson DT, Xu J, Cherry JH, Urry DW, Daniell H (1995) Hyper expression of an environmentally friendly synthetic polymer gene. Biotechnol Lett 17:745–750
Guda C, Lee SB, Daniell H (2000) Stable expression of a biodegradable protein-based polymer in tobacco chloroplasts. Plant Cell Rep 19:257
Herzog RW, Singh NK, Urry DW, Daniell H (1997) Expression of a synthetic protein-based polymer (elastomer) gene in Aspergillus nidulans. Appl Microbiol Biotechnol 47:368–372
Heslot H (1998) Artificial fibrous proteins: a review. Biochimie 80:19–31
Inan M, Chiruvolu V, Eskridge KM, Vlasuk GP, Dickerson K, Brown S, Meagher MM (1999) Optimization of temperature-glycerol-pH conditions for a fed-batch fermentation process for recombinant hookworm (Ancylostoma caninum) anticoagulant peptide (AcAP-5) production by Pichia pastoris. Enzyme Microb Technol 24:438–445
Jahic M, Gustavsson M, Jansen AK, Martinelle M, Enfors SO (2003) Analysis and control of proteolysis of a fusion protein in Pichia pastoris fed-batch processes. J Biotechnol 102:45–53. doi:https://doi.org/10.1016/S0168-1656(03)00003-8
Kreil G (1990) Processing of precursors by dipeptidylaminopeptidases: a case of molecular ticketing. Trends Biochem Sci 15:23
Lee C, Levin A, Branton D (1987) Copper staining: a five-minute protein stain for sodium dodecyl sulfate-polyacrylamide gels. Anal Biochem 166:308
Li PZ, Anumanthan A, Gao XG, Ilangovan K, Suzara VV, Duzgunes N, Renugopalakrishnan V (2007) Expression of recombinant proteins in Pichia pastoris. Appl Biochem Biotechnol 142:105–124
Matoba S, Fukayama J, Wing RA, Ogrydziak DM (1988) Intracellular precursors and secretion of alkaline extracellular protease of Yarrowia lipolytica. Mol Cell Biol 8:4904–4916
Mattanovich D, Gasser B, Hohenblum H, Sauer M (2004) Stress in recombinant protein producing yeasts. J Biotechnol 113:121–135
McHale MK, Setton LA, Chilkoti A (2005) Synthesis and in vitro evaluation of enzymatically cross-linked elastin-like polypeptide gels for cartilaginous tissue repair. Tissue Eng 11:1768–1779
Megeed Z, Cappello J, Ghandehari H (2002) Genetically engineered silk-elastinlike protein polymers for controlled drug delivery. Adv Drug Deliv Rev 54:1075
Meyer DE, Chilkoti A (1999) Purification of recombinant proteins by fusion with thermally-responsive polypeptides. Nat Biotechnol 17:1112–1115
Meyer DE, Chilkoti A (2002) Genetically encoded synthesis of protein-based polymers with precisely specified molecular weight and sequence by recursive directional ligation: examples from the elastin-like polypeptide system. Biomacromolecules 3:357–367. doi:https://doi.org/10.1021/bm015630n
Meyer DE, Chilkoti A (2004) Quantification of the effects of chain length and concentration on the thermal behavior of elastin-like polypeptides. Biomacromolecules 5:846–851
Mithieux SM, Weiss AS (2005) Elastin. In: Parry AD (ed) Fibrous proteins: coiled-coils, collagen and elastomers. Elsevier Academic, San Diego, pp 437–458
Rincon AC, Molina-Martinez IT, de Las Heras B, Alonso M, Bailez C, Rodriguez-Cabello JC, Herrero-Vanrell R (2006) Biocompatibility of elastin-like polymer poly(VPAGV) microparticles: in vitro and in vivo studies. J Biomed Mat Res Part A 78A:343–351. doi:https://doi.org/10.1002/jbm.a.30702
Rodriguez-Cabello JC, Prieto S, Reguera J, Arias FJ, Ribeiro A (2007) Biofunctional design of elastin-like polymers for advanced applications in nanobiotechnology. J Biomater Sci, Polym Ed 18:269–286. doi:https://doi.org/10.1163/156856207779996904
Simnick AJ, Lim DW, Chow D, Chilkoti A (2007) Biomedical and biotechnological applications of elastin-like polypeptides. Polym Rev 47:121–154
Sinha J, Plantz BA, Inan M, Meagher MM (2005) Causes of proteolytic degradation of secreted recombinant proteins produced in methylotrophic yeast Pichia pastoris: case study with recombinant ovine interferon-τ. Biotechnol Bioeng 89:102–112. doi:https://doi.org/10.1002/bit.20318
Toonkool P, Weiss AS (2001) Expression of recombinant human tropoelastin in Saccharomyces cerevisiae containing a synthetic gene with a high codon adaptation index coupled to the SUC2 invertase signal sequence. Acta Biotechnol 21:189–193
Trabbic-Carlson K, Setton LA, Chilkoti A (2003) Swelling and mechanical behaviors of chemically cross-linked hydrogels of elastin-like polypeptides. Biomacromolecules 4:572–580. doi:https://doi.org/10.1021/bm025671z
Trabbic-Carlson K, Liu L, Kim B, Chilkoti A (2004) Expression and purification of recombinant proteins from Escherichia coli: comparison of an elastin-like polypeptide fusion with an oligohistidine fusion. Protein Sci 13:3274–3284
Urry DW (1976) Molecular mechanisms of elastin coacervation and coacervate calcification. Faraday Discus 205–212
Urry DW (1992) Free energy transduction in polypeptides and proteins based on inverse temperature transitions. Prog Biophys Mol Biol 57:23–57
Urry DW, Long MM, Cox BA, Ohnishi T, Mitchell LW, Jacobs M (1974) Synthetic polypentapeptide of elastin coacervates and forms filamentous aggregates. Biochim Biophys Acta, Protein Struct 371:597–602
Urry DW, Luan CH, Parker TM, Gowda DC, Prasad KU, Reid MC, Safavy A (1991a) Temperature of polypeptide inverse temperature transition depends on mean residue hydrophobicity. J Am Chem Soc 113:4346–4348
Urry DW, Parker TM, Reid MC, Gowda DC (1991b) Biocompatibility of the bioelastic materials, poly(GVGVP) and its gamma-irradiation cross-linked matrix—summary of generic biological test-results. J Bioact Compat Polym 6:263–282
Urry DW, Gowda DC, Parker TM, Luan CH, Reid MC, Harris CM, Pattanaik A, Harris RD (1992) Hydrophobicity scale for proteins based on inverse temperature transitions. Biopolymers 32:1243–1250
Wenzel T, Sparbier K, Mieruch T, Kostrzewa M (2006) 2, 5-dihydroxyacetophenone: a matrix for highly sensitive matrix-assisted laser desorption/ionization time-of-flight mass spectrometric analysis of proteins using manual and automated preparation techniques. Rapid Commun Mass Spectrom 20:785–789. doi:https://doi.org/10.1002/rcm.2378
Werten MWT, Van den Bosch TJ, Wind RD, Mooibroek H, De Wolf FA (1999) High-yield secretion of recombinant gelatins by Pichia pastoris. Yeast 15:1087–1096
Werten MWT, Wisselink WH, Jansen-van den Bosch TJ, de Bruin EC, de Wolf FA (2001) Secreted production of a custom-designed, highly hydrophilic gelatin in Pichia pastoris. Protein Eng 14:447–454
Werten MWT, Moers APHA, Vong T, Zuilhof H, van Hest JCM, de Wolf FA (2008) Biosynthesis of an amphiphilic silk-like polymer. Biomacromolecules 9:1705–1711
Zhang X, Guda C, Datta R, Dute R, Urry DW, Daniell H (1995) Nuclear expression of an environmentally friendly synthetic protein based polymer gene in tobacco cells. Biotechnol Lett 17:1279–1284
Zhang W, Bevins M, Plantz B, Smith L, Meagher M (2000) Modeling Pichia pastoris growth on methanol and optimizing the production of a recombinant protein, the heavy-chain fragment C of botulinum neurotoxin, serotype A. Biotechnol Bioeng 70:1–8
Zhang W, Inan M, Meagher MM (2007) Rational design and optimization of fed-batch and continuous fermentations. In: Cregg J (ed) Pichia protocols, 3rd edn. Humana, Totowa, pp 43–63
Zhao W, Wang JW, Deng RQ, Wang XZ (2008) Scale-up fermentation of recombinant Candida rugosa lipase expressed in Pichia pastoris using the GAP promoter. J Ind Microbiol Biotech 35:189–195
Acknowledgment
The authors are grateful to Aernout Martens who provided the pMTL23-AIII cloning vector that was used for oligomerization and Antoine Moers for his assistance in the practical work. This project is financially supported by the Netherlands Ministry of Economic Affairs and the B-Basic partner organizations (www.b-basic.nl) through B-Basic, a public–private NWO-ACTS program (ACTS = Advanced Chemical Technologies for Sustainability).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Schipperus, R., Teeuwen, R.L.M., Werten, M.W.T. et al. Secreted production of an elastin-like polypeptide by Pichia pastoris . Appl Microbiol Biotechnol 85, 293–301 (2009). https://doi.org/10.1007/s00253-009-2082-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-009-2082-9