Abstract
Mapping resistance genes is usually accomplished by phenotyping a segregating population for the resistance trait and genotyping it using a large number of markers. Most resistance genes are of the NBS-LRR type, of which an increasing number is sequenced. These genes and their analogs (RGAs) are often organized in clusters. Clusters tend to be rather homogenous, viz. containing genes that show high sequence similarity with each other. From many of these clusters the map position is known. In this study we present and test a novel method to quickly identify to which cluster a new resistance gene belongs and to produce markers that can be used for introgression breeding. We used NBS profiling to identify markers in bulked DNA samples prepared from resistant and susceptible genotypes of small segregating populations. Markers co-segregating with resistance can be tested on individual plants and directly used for breeding. To identify the resistance gene cluster a gene belongs to, the fragments were sequenced and the sequences analyzed using bioinformatics tools. Putative map positions arising from this analysis were validated using markers mapped in the segregating population. The versatility of the approach is demonstrated with a number of populations derived from wild Solanum species segregating for P. infestans resistance. Newly identified P. infestans resistance genes originating from S. verrucosum, S. schenckii, and S. capsicibaccatum could be mapped to potato chromosomes 6, 4, and 11, respectively.
Similar content being viewed by others
Introduction
Plants are attacked by a wide range of pathogens including viruses, bacteria, oomycetes, fungi, nematodes, and insects. They have evolved passive and active ways to defend themselves against these attackers. One of the active defense systems is a type of immunity that is described by the gene for gene resistance theory, which was developed by Flor in the 1940s. It considers the gene causing resistance, the R gene in the host, to be complementary to an Avr (avirulence) gene in the pathogen (Flor 1942, 1971). To date, more than 90 resistance (R) genes have been identified in various plants, by a wide variety of methods including map-based cloning, transposon tagging, and similarity-based DNA library screening (Ingvardsen et al. 2008). Most R genes can be assigned to one of the five major classes of R genes (Dangl and Jones 2001). The largest of these classes contain genes that encode proteins with a nucleotide binding site and a leucine-rich repeat region (the so called NBS-LRR genes). NBS-LRR resistance genes and their analogs (RGAs) are numerous in plant genomes and are often organized in clusters (Michelmore and Meyers 1998; AGI 2000). Many of the R genes in Solanum seem to be positioned in relatively few clusters (Gebhardt et al. 1991; Tanksley et al. 1992; Bakker et al. 2003).
The common approach to map resistance genes is to construct a mapping population derived from a susceptible and a resistant parent, phenotype the offspring, and then analyze the offspring with molecular markers. As many resistance traits turned out to be controlled by a single gene, more efficient methods have been developed to facilitate the search for markers linked to these genes. Bulked segregant analysis is a method for efficiently identifying markers linked to a specific trait. Two pooled DNA samples of individuals from a segregating population with contrasting phenotypes resulting from a single cross are compared. Michelmore et al. (1991) showed that this approach works well to rapidly identify RAPD and RFLP markers linked to the trait of interest.
The NBS region of (NBS-LRR) R genes and RGAs contains highly conserved common motifs like the P-loop, the kinase-2 motif, and the GLPL motif (Meyers et al. 1999, 2003; Monosi et al. 2004). These conserved motifs within the NBS-LRR genes have been used successfully to sequence parts of NBS regions from various plant species (Collins et al. 1998; Pflieger et al. 1999; Zhang et al. 2007). Van der Linden et al. (2004) developed a method for efficiently tagging NBS-LRR type of resistance genes and their analogs called NBS profiling. NBS profiling is a PCR-based method that makes use of primers that target different conserved motifs in the NBS domain. It produces a DNA profile that is highly enriched for R genes and RGAs. Studies in apple (Calenge et al. 2005) and in potato, tomato, barley, and lettuce (van der Linden et al. 2004) show that NBS profiling produces markers that are tightly linked to R genes and R-gene clusters.
Late blight caused by the oomycete Phytophthora infestans is one of the most important and devastating diseases in potato. Currently, late blight is mainly controlled by a combination of disease management strategies, relying heavily on the use of fungicides (Fry 2008). High disease management costs, environmental concern, and the threat of promoting the evolution of resistant populations stimulated the search for R genes that can be used in breeding programs to create resistant cultivars. In the past, 11 late blight resistance genes from the wild potato species Solanum demissum (Gebhardt and Valkonen 2001) were introduced into cultivated potato. As some of the resistances conferred by these R genes were quickly overcome by the pathogen (Wastie 1991) the focus of breeders and scientist moved toward germplasm with partial or quantitative resistance (van der Vossen et al. 2005; Fry 2008). More recently, the interest in finding new R genes has increased again. The presence of R genes conferring resistance against P. infestans in other wild potato species than S. demissum was investigated as well. Resistance against P. infestans conferred by R genes has been found in S. pinnatisectum (Kuhl et al. 2001), S. bulbocastanum (Naess et al. 2000; Song et al. 2003; van der Vossen et al. 2003, 2005; Park et al. 2005a), S. berthaultii (Ewing et al. 2000; Rauscher et al. 2006), S. microdontum (Sandbrink et al. 2000; Tan et al. 2008), S. mochiquense (Smilde et al. 2005), S. paucissectum (Villamon et al. 2005), S. phureja (Śliwka et al 2006) S. venturii (Pel et al. 2009, Foster et al. 2009), and S. stoloniferum (Wang et al. 2008a).
There are still many other wild species that have not been tested yet for the presence of R genes against P. infestans. In the present study, we searched for new P. infestans R genes and markers in the wild potato species S. verrucosum, S. schenckii, and S. capsicibaccatum. We present and test a novel approach to quickly identify at which chromosome/chromosomal region the targeted resistance gene is located and to obtain markers that can be used for introgression breeding. The novelty of the approach is in the combination of small size segregating populations in combination with NBS profiling techniques and bioinformatics to quickly retrieve a putative map position of the segregating gene. We illustrate this approach by describing three cases using wild Solanum populations that are segregating for P. infestans resistance.
Materials and methods
Plant material
The plant material used as parents for the segregating populations was selected from a large screen of around 1,000 accessions of mainly wild Solanum section Petota germplasm. The evaluated material was described by Jacobs et al. (2008). The following segregating populations were used in this study and details on the progeny can be found in Table 1. The ver03-392 and ver03-394 are very similar BC2 populations, both S. verrucosum derived mothers (ver00-3228 and ver00-3229) are late blight resistant clones derived from the same cross (ver 98-28-2 × AR 93-2090) and they were crossed with the same susceptible male parent. The ver 98-28-2 clone is derived from CGN 17772 (BGRC8255). The population snk7458 was a F1 population based on a resistant individual S. schenckii GLKS 30659 crossed with a sensitive parent S. brachycarpum CGN 18347. Population cap7358 was a F1 population derived from a cross with resistant parent S. capsicibaccatum CGN 22388 and susceptible parent S. circaeifolium CGN 18133.
Phytophthora infestans isolate and disease testing
The aggressive and complex P. infestans isolate 90128 (race 1.3.4.7.8.11), kindly provided by Prof. Francine Govers (Laboratory of Phytopathology, Wageningen University) was cultured on cultivar Bintje leaves or on rye sucrose medium as described previously (Vleeshouwers et al. 1999). For disease testing, leaves from 8 to 10-week-old plants were used. The third and fourth fully stretched leaves (counted from the top) were detached, and placed in water-saturated florists foam. For each plant genotype, two replicates were inoculated on five leaflets of one leaf, resulting in 10 replicates per genotype. The leaves were inoculated with a zoospore suspension of 50,000 spores/ml and incubated in humid trays. After 6 days, the leaves were examined for occurrence of sporulation and the lesions sizes (LS) measured. The measurements were normalized for the lesion size using the reference cultivar Bintje.
DNA extractions and NBS profiling
After 7 to 8 weeks of growing, young plant leaves were harvested for DNA extraction. DNA was extracted according to Fulton et al. (1995). NBS profiling was performed as described by van der Linden et al. (2004), with some minor modifications. The protocol of NBS profiling involves three steps: (1) restriction enzyme digestion of genomic DNA and the ligation of adapters (which in our experiments was done in one and the same incubation step, except when the enzyme TaqI was used), (2) selective amplification of fragments containing an NBS motif using a degenerated primer for the conserved domains, and (3) polyacrylamide gel electrophoresis of the amplified fragments.
The following 5 restriction enzymes: MseI, TaqI, RsaI, AluI, and HaeIII, were used in combination with 5 NBS primers: NBS1, NBS2, NBS3, NBS5a6, and NBS9 (van der Linden et al. 2004; Wang et al. 2008b; Mantovani et al. 2006; Brugmans et al. 2008), resulting in 25 primer–enzyme combinations. NBS profiling was carried out first on the parents and on bulks of pooled resistant and susceptible plants from the population of interest. When this first round produced polymorphic bands between the parents and between the bulks, another round of NBS profiling was carried out on the DNA of the parents, the bulks, and all individuals separately that constituted the resistant and susceptible bulk using only the primer–enzymes combinations that produced polymorphic bands. In the two small S. verrucosum populations ver03-392 and ver03-394, only samples from 5 resistant and 3 susceptible and 7 resistant and 5 susceptible plants, respectively, could be scored reliably for P. infestans resistance.
Sequence analysis of polymorphic bands
To determine the sequence of a polymorphic NBS marker, the band was excised from the gel and re-amplified with the same primers that initially produced the band. The PCR conditions were identical to the first PCR of the NBS profiling protocol. Only bands that were clearly separated from surrounding bands were considered. In the case of population 7358 it was necessary to clone the band first because direct sequencing showed that the band consisted of a mixture of two fragments. For this, the PCR products were ligated into the pGEM-T easy Vector System (Promega). Ligation mixtures were transformed into E. coli DH5α, as recommended by the supplier (Invitrogen). Colonies containing a plasmid with insert were used for colony PCR. Clones were sequenced using vector M13 primers. For populations ver03-392, ver03-394, and snk7458 the bands were directly sequenced following reamplification. Each fragment was sequenced from both sides with the NBS profiling primers using the Big Dye Terminator Kit on an ABI 3700 automated sequencer (Applied Biosystems, USA). DNA sequences were analyzed using DNAstar (Lasergene, Madison, WI, USA). The obtained sequences were compared to the NCBI nucleotide database (http://www.ncbi.nlm.nih.gov/), by using the discontiguous megablast option of the BLASTN suite (Altschul et al. 1997) from NCBI. The similarity scores with sequences found in the NCBI database were evaluated taking into account the E value. The E value is also dependent on the length of the query sequence that can be blasted to a certain sequence in the database. The shorter the sequence, the higher the possibility that the result is due by chance. A similarity was defined as a good hit if it showed a combination of a similarity (identity) score of 75% or higher plus a small expected value of 1.00E-25 or smaller. The sequences have been submitted to Genbank and are available under the accession numbers GU060647–GU060661 (see also Table 3).
Confirmation of position with PCR-based primers
To verify the putative map positions for NBS profiling markers deduced from the BLAST analysis, we used flanking markers (mainly CAPS). For each population, flanking markers were tested on the parents and all individuals of the cross that could be scored as clearly resistant or susceptible. Many CAPS and SCAR markers were tested, but only few showed polymorphisms in the offspring. The details on the primers that were successfully used are given in Table 4. To amplify the samples with PCR marker Th21, approximately 10 ng of genomic DNA was mixed in a total volume of 20 μl containing (final concentration per reaction) 1× PCR buffer, 0.2 mM mixture of all dNTPs, 0.1 pm of each primer and 0.1 unit Taq DNA polymerase (Promega). The following PCR protocol was used: a first step of 3 min at 96°C, followed by 30 cycles of 0.5 min at 96°C, 0.5 min at 56°C, and 1 min at 72°C, concluding with 10 min at 72°C. With the CAPS markers CP58 and CD67 a slightly different mixture and protocol was used. Approximately 10 ng of DNA was mixed in a total volume of 25 μl containing 1× PCR buffer, 0.12 mM dNTPs, 0.05 pM from each primer, and 0.1 unit Super Taq DNA polymerase. The following PCR protocol was used: starting with 4 min at 94°C, followed by 35 cycles of 0.5 min at 94°C, 0.5 min of 58°C, and 1.5 min at 72°C. At the end of the protocol, 6 min at 72°C were programmed. The presence of PCR products of the correct length was evaluated on a 1.0% agarose gel.
Results
Segregation analysis for resistance to P. infestans
To analyze whether R genes segregate in the generated populations, the offspring and parents were inoculated with P. infestans and assessed for resistance. From the ver03-392 and ver03-394 populations only very few progeny plants were available. Still, for both populations a segregation for resistance could be detected in repeated experiments, although not all individuals could be scored as clearly resistant or susceptible (Table 1). Population snk7458 also segregated for resistance to P. infestans isolate 90128 yet there were far more resistant that susceptible individuals, i.e. 39 and 5, respectively, and in addition a few genotypes with intermediate phenotypes. For cap7358, a clear segregation between resistance and susceptibility was evident, and no intermediate phenotypes were observed. From all populations, subsets of genotypes with reliable phenotypes could now be assembled as resistant and susceptible bulks (Table 1), for further analysis by NBS profiling.
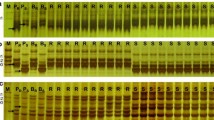
NBS profiling
The populations differed strongly in the number of polymorphic bands in the bulks that showed co-segregation with resistance, ranging from 1 in population cap7358 to 33 in the ver03-392 population (Table 2). For populations snk7458, cap7358, and ver03-394 all the primer/enzyme combinations that produced polymorphic bands were tested on the individuals of the bulks. In the population ver03-392, only a selection of primer/enzyme combinations producing polymorphic bands in the bulks was tested on the individuals. An example of an NBS gel for parents, bulks, and individuals is given in Fig. 1. Not all putative polymorphisms observed in the bulks were validated in the individuals (see Table 2). Bands that were found and confirmed as co-segregating in the second round of NBS profiling on individuals of the bulks were bands in coupling phase, e.g. co-segregating with the resistant phenotype, or in repulsion phase, e.g. co-segregating with the susceptible phenotype. All NBS markers that were studied in the individuals of the bulks co-segregated with resistance or susceptibility as expected, except for the NBS markers ver03-394_9H1, ver03-394_9R1, and ver03-394_9R2 which show 1 (identical) recombinant resistant plant (out of 7 resistant plants) that does not have the specific NBS fragment.
Identification of NBS bands and deduction of map positions
The bands co-segregating with the resistance phenotype in the individuals were excised from the gels and sequenced to determine their identity. Sequences obtained were compared to the NCBI database using BLAST analysis. Most of these sequences gave hits with NBS-related sequences, so these sequences we regarded as RGAs. For a few sequences there were no hits at all with NCBI database. The best hits (high identity score and low E value) of these sequences to Solanum sequences are shown in Table 3.
For population ver03-392, 2 bands were successfully sequenced, but only band 392-9H1, see Table 3, gave high similarity scores, higher than 75%, with Solanum sequences from Genbank. Good hits were found with S. lycopersicum chromosome 11 clone C11HBa0119D16, complete sequence, and later with S. lycopersicum DNA, chromosome 8, clone: C08SLm0114A09, complete sequence. For population ver03-394, 5 different NBS bands could be sequenced successfully, 4 bands gave high similarity scores with Solanum sequences from Genbank. One of them (394_9H1) was identical to the 392-9H1 band. The bands 394_H1, 394_R1, and 394_R2 are identical in the overlapping region. The bands 394_H1 and 394_T1 show a 98% similarity in the overlapping region. The sequences found in the NCBI database that showed similarity with the NBS markers of ver03-394 were identical to those found for population ver03-392.
For population snk7458, sequences of 5 NBS profiling bands could be successfully retrieved. Among each other the fragments show between 70 and 92% similarity in the overlapping regions. For all 5 bands, the highest similarity was found with S. lycopersicum BAC clone Clemson_Id 127E11. Park et al. (2005a) showed that this BAC clone contains several RGA sequences that are similar to the Rpi-blb3 gene of S. bulbocastanum which is located on chromosome 4 of potato (Park et al. 2005a). Another high similarity score with bands from snk7458 was found with a S. lycopersicum DNA sequence from clone SL_MboI-40B16, also located on chromosome 4.
For population cap7358, the only band, cap7358_3M1, found in the NBS profiling analysis with the bulk and the individual samples was sequenced. After direct sequencing, it appeared that the band was a mixture between at least two fragments. The band was, therefore, cloned and sequenced. As much as 5 out of 10 cloned sequences were almost identical, 2 others were almost identical to each other but different from the others, and the last 3 were different from all. The sequence of the five identical fragments gave, when compared with the sequences in the Genbank database, an identity score of 91% with an E value of 7.00E-27 (Table 3) with S. tuberosum mRNA for the NL27 protein. Hehl et al. (1999) located the gene encoding the NL27 protein on chromosome 11.
Confirmation of map position of genes
To verify the deduced map positions, we used markers that were expected to be (closely) linked based on their position on the potato maps (http://www.gabipd.org/database/maps.html and http://www.sgn.cornell.edu/). For each population, flanking markers were tested on the parents and all offspring that could be scored as clearly resistant or susceptible. The results are presented in Table 4. For populations ver03-392 and ver03-394 sequence similarity suggested that the resistance gene was positioned on chromosome 11. Several CAPS markers for chromosome 11 were tested, but none of those displayed any polymorphisms. The marker sequence was compared to a sequence database containing NBS profiling marker sequences that were mapped in the SHxRH potato mapping population (van der Linden et al., unpublished results), and was found to be nearly identical to a marker mapped on chromosome 6. This mapping position was confirmed by marker CD67 digested with enzyme HpyCH4IV and with enzyme SsiI that both produced a polymorphic band that co-segregated with the resistance (see Fig. 2). An extra band is visible in the resistant parent and the resistant offspring in both S. verrucosum populations.
The P. infestans resistance in population snk7458 was suggested to be located in the same cluster as the Rpi-blb3 gene on chromosome 4. Several markers for chromosome 4, such as TG370, Th21, TG506R, CT229, and T1430, were tested. The parents often showed polymorphisms, but the offspring was almost always homogeneous for the same marker. Therefore, another approach was taken to find segregating markers. The PCR products of markers TG506, AF411807R, T1430, TG370, and Th21 from 6 resistant and 5 susceptible individuals were sequenced and checked for SNPs. The only polymorphism that was found between the resistant and susceptible individuals was in the PCR product of marker Th21. This SNP was shown to co-segregate with resistance. The SNP is located in an MboI site in the middle of the PCR fragment (TGATC for the susceptible, T[A/G]ATC for the (heterozygous) resistant individuals). A PCR with Th21 followed by digestion with MboI on all the available individuals, resulted in an extra band for the resistant parent S. schenckii GLKS 30659 and an extra band for 27 out of 39 resistance phenotypes. All the 5 susceptible phenotypes lacked the extra band. This implies that at least 12 out of 45 resistant plants contain a second gene conferring resistant to P. infestans.
To confirm the position of the marker in population cap7358, several SSR and CAPS primers for chromosome 11 were tested on the parents, the bulks and the individuals of this population. The CAPS marker CP58 in combination with restriction enzyme MspI produced an extra band in the resistant parent and in 18 out of 18 susceptible offspring, showing that the band was segregating in repulsion phase. Thirty one out of the 32 resistant individuals lacked this extra band.
Discussion
A new strategy for mapping resistance genes
In the present study we describe a novel approach to identify markers linked to resistance genes. The first step in the approach consists of producing small populations segregating for P. infestans resistance, phenotyping the populations for resistance, and composing bulks of resistant and susceptible individuals. Then, the bulks are genotyped using NBS profiling to obtain markers that co-segregate with resistance, followed by sequencing of co-segregating NBS fragments and BLAST analysis to identify the fragment. Combining this information with literature data on mapping of resistance genes results in a suggestion for a putative map position. Finally, the map positions are confirmed using known flanking markers.
Large differences were observed in the number of markers co-segregating with resistance in the 3 different populations, ranging from 1 in population cap7358 to 33 in the ver03-392 population (Table 2). These differences are possibly caused by the position of the targeted R gene. A high number of polymorphic bands co-segregating with resistance probably means that they are part of a large cluster of R genes. Few or no polymorphic NBS bands could mean that the parents were closely related and that the targeted R gene has an isolated position. In case no polymorphisms are detected with the 25 primer–enzyme combinations additional enzymes or primers may be tested. Furthermore, there is always a chance that the resistance gene under study is not of the NBS-LRR type but belongs to another class of resistance genes (Ingvardsen et al. 2008). The polymorphisms identified in the bulks that could not be confirmed in the analysis of the individual genotypes were due to differences in band intensity. The polymorphisms that showed a clear presence/absence of bands could be confirmed.
Many NBS profiling bands that were sequenced could be annotated successfully as being from a putative NBS-LRR type of resistance gene (results not shown). Correct annotation depends on the availability of sufficient sequence information in the databases. In addition, highly homologous sequences may sometimes be found in different clusters, as was shown for Mi and I2 homologs (van der Vossen et al. 2005; Seah et al. 2007). This may complicate mapping afterwards, as was shown for fragment 392_9H1, obtained from population ver392. For this fragment, high similarity was found with a sequence of S. lycopersicum which had previously been mapped to chromosome 11 (Mueller et al. 2005, 2008), while another sequence homolog was retrieved from chromosome 4 (McGuire 2009). The putative positions on chromosome 11, or chromosomes 8 or 4, (Table 3), as suggested by BLAST with the NCBI database for several bands from the populations ver03-392 to ver03-394, could not be verified. The putative map position on chromosome 6 for this marker, inferred from the high sequence similarity to a mapped NBS profiling marker (van der Linden et al., unpublished results) could be verified. This indicates that the NCBI database is still far from complete. With the increasing amount of data in public sequence databases and with the progress of the potato genome sequencing (PGSC, http://www.potatogenome.net) it is likely that in the near future new fragments can be mapped more efficiently with higher accuracy.
Identification and mapping of P. infestans resistance genes in S. verrucosum, S. schenckii, and S. capsicibaccatum
In the ver03-392 and ver03-394 populations, the resistance against P. infestans is located on chromosome 6, near marker CD67. Population 03-394 showed one recombinant for the polymorphism found with marker CD67. We have named the gene underlying the resistance Rpi-ver1. As we could not find any other co-segregating markers for the resistance, it is not clear whether the gene is positioned downstream or upstream of marker CD67. The marker CD67 itself is positioned at 10.5 cM according to the potato map Potato-TXB 1992 v27 (http://www.sgn.cornell.edu/). Resistance against P. infestans in S. verrucosum has been reported by van Soest et al. (1984). Rivera-Peña and Molina-Galan (1989) and Rivera-Peña (1990) studied the occurrence of late blight on naturally occurring populations of wild Solanum species on the slope of Nevado de Toluca for many years. He found highly resistant natural populations of S. verrucosum. Furthermore, a P. infestans resistance screening of the Commonwealth potato collection also yielded a very resistant S. verrucosum accession (Bradshaw et al. 2006). Whether any of the genes involved is similar to the Rpi-ver1 remains to be established. Finally, Liu and Halterman (2006) reported on P. infestans resistance in S. verrucosum. They have identified a gene sharing 83.5% nucleotide identity with Rpi-blb1. It will be interesting to see whether this gene maps to the same position on chromosome 8 as the original Rpi-blb1 (van der Vossen et al. 2003) or to the same position as Rpi-ver1 on chromosome 6, or to a complete new position.
There are no indications that the sequences of the NBS markers of populations ver03-392 and ver03-394 show any similarity to the original Rpi-blb1.
In the snk7458 population, there is probably more than 1 resistance gene against P. infestans segregating, since the offspring consists of far more resistant than susceptible phenotypes (39:5). Furthermore, only 2/3 of the resistant genotypes contained the linked CAPS marker. None of the susceptible phenotypes contained the CAPS marker. This result suggests the possible presence of another gene or QTL that confers the phenotypic resistance. This situation is very similar to that described by Wang et al. (2008a) for S. stoloniferum. Based on the results, one would also expect the presence of polymorphic NBS bands with <100% co-segregation. However, the five NBS markers tested on 6 resistance and 4 susceptible individuals from the bulk showed 100% co-segregation for the NBS markers. It is possible that the polymorphic NBS bands that were linked to the other gene were not discovered in the first round of NBS profiling on the bulked individuals. One gene conferring resistance in the S. schenckii population snk7458, and mapped in this study is most likely located on chromosome 4, near or on marker Th21 (Table 4). We call the gene underlying the resistance Rpi-snk1. Our results indicate that Rpi-snk1 is a member of a cluster of Rpi-blb3 homologs on chromosome 4 (Park et al. 2005b). It co-segregates with the Th21 marker, and therefore it is also similar to Rpi-abpt, R2, and R2-like that all reside in the same R gene cluster on chromosome 4 (Park et al. 2005b). According to the phenetic and phylogenetic results of Jacobs et al. (2008), S. schenckii is closely related to S. hougasii. Accessions from S. hougasii are reported to show high resistance against P. infestans (Bradshaw et al. 2006). It will be interesting to see whether these accessions also carry the same resistance genes as found in S. schenckii.
In population cap7358, a gene conferring resistance against P. infestans was found on chromosome 11, near marker CP58. The marker identified is in repulsion phase. In the 50 offsprings tested with the CP58 marker only one individual had lost linkage with this marker. The newly found gene is named Rpi-cap1. The position of marker CP58 is at the top of the chromosome 11, on 0.00 cM according to the data from the map Potato-TXB 1992 V27 (http://www.sgn.cornell.edu/).
Another resistance gene that was mapped to this region is R-mc1 (mapped at 66 cM in the functional map of chromosome 11 of potato for pathogen resistance, as published at http://www.gabipd.org/database/maps.shtml), which is a resistance gene against root-knot nematode Meloidogyne chitwoodi retrieved from S. bulbocastanum (Brown et al. 1996). Note that the orientation of this potato map of chromosome 11 is reversed compared to the previously mentioned SGN potato map of chromosome 11. Resistance against P. infestans in S. capsicibaccatum was reported by van Soest et al. (1984) and Ruiz de Galarreta et al. (1998) but no further details on sequence or position of this R gene are available.
Very recently, Pel et al. (2009) described the cloning of two P. infestans resistance genes from S. venturii. The genes were mapped to chromosome 9 using NBS profiling. The NBS profiling fragments were sequenced and identified as a Tm-2 2 homologs. They designed degenerated primers based on all known Tm-2 2, which enabled them to clone the genes involved. This example shows the versatility of NBS profiling in combination with an allele mining strategy for quickly identifying and cloning of resistance genes.
In the mapping populations used in this study no new resistance gene clusters in Solanum material were found. The P. infestans resistance was derived from different wild Solanum species in which previously no resistance genes had been identified (though some species had been reported to express some P. infestans resistance). Although we used these relatively unknown sources, it seems that the genes conferring the resistance are linked to known clusters of resistance genes. This may suggest that the present view on the Solanum genome is rather exhaustive and that most resistance clusters are already known. In a previous study, new resistance genes in Solanum derived from wild Solanum species, could also be positioned at already known R gene clusters of the genome. Wang et al. (2008a) found that the dominant R genes Rpi-sto1 (derived from S. stoloniferum) and Rpi-plt1 (from S. polytrichon) resided at the same position on chromosome 8 as Rpi-blb1 in S. bulbocastanum. Possibly the found R genes on known loci constitute new alleles but new alleles can be positively identified with the aid of effector proteins (Vleeshouwers et al. 2008).
References
AGI (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408:796–815
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Bakker E, Butterbach P, van der Rouppe Voort J, van der Vossen E, van Vliet J, Bakker J, Goverse A (2003) Genetic and physical mapping of homologues of the virus resistance gene Rx1 and the cyst nematode resistance gene Gpa2 in potato. Theor Appl Genet 106:1524–1531
Bradshaw J, Bryan G, Ramsay G (2006) Genetic resources (including wild and cultivated Solanum species) and progress in their utilisation in potato breeding. Potato Res 49:49–65
Brown CR, Yang C-P, Mojtahedi H, Santo GS, Masuelli R (1996) RFLP analysis of resistance to Columbia root-knot nematode derived from Solanum bulbocastanum in a BC2 population. Theor Appl Genet 92:572–576
Brugmans B, Wouters D, van Os H, Hutten R, van der Linden G, Visser R, van Eck H, van der Vossen E (2008) Genetic mapping and transcription analyses of resistance gene loci in potato using NBS profiling. Theor Appl Genet 117:1379–1388
Calenge F, van der Linden CG, van de Weg E, Schouten HJ, van Arkel G, Denancé C, Durel CE (2005) Resistance gene analogues identified through the NBS-profiling method map close to major genes and QTL for disease resistance in apple. Theor Appl Genet 110:660–668
Collins NC, Webb CA, Seah S, Ellis JG, Hulbert SH, Pryor A (1998) The isolation and mapping of disease resistance gene analogs in maize. Mol Plant Microbe Interact 11:968–978
Dangl JL, Jones CJ (2001) Plant pathogens and integrated defence responses to infection. Nature 411:826–833
Ewing EE, Simko I, Smart CD, Bonierbale MW, Mizubuti ESG, May GD, Fry WE (2000) Genetic mapping from field tests of qualitative and quantitative resistance to Phytophthora infestans in a population derived from Solanum tuberosum and Solanum berthaultii. Mol Breed 6:25–36
Flor HH (1942) Inheritance of pathogenicity in Melampsora lini. Phytopathology 32:653–669
Flor HH (1971) The current status of the gene for gene concept. Ann Rev Phytopathol 9:275–296
Foster SJ, Park T-H, Pel MA, Brigneti G, Śliwka J, Jagger L, Van der Vossen EAG, Jones JDG (2009) Rpi-vnt1.1, a Tm-22 homolog from Solanum venturii confers resistance to potato late blight. Mol Plant Microbe Interact 22:589–600
Fry W (2008) Phytophthora infestans: the plant (and R gene) destroyer. Mol Plant Path 9:385–402
Fulton T, Chunwongse J, Tanksley S (1995) Microprep protocol for extraction of DNA from tomato and other herbaceous plants. Plant Mol Biol Rep 13:207–209
Gebhardt C, Valkonen JPT (2001) Organization of genes controlling disease resistance in the potato genome. Ann Rev Phytopathol 39:79–102
Gebhardt C, Ritter E, Barone A, Debener T, Walkemeier B, Schachtschabel U, Kaufmann H, Thompson RD, Bonierbale MW, Ganal MW, Tanksley SD, Salamini F (1991) RFLP maps of potato and their alignment with the homoeologous tomato genome. Theor Appl Genet 83:49–57
Hehl R, Faurie E, Hesselbach J, Salamini F, Whitham S, Baker B, Gebhardt C (1999) TMV resistance gene N homologues are linked to Synchytrium endobioticum resistance in potato. Theor Appl Genet 98:379–386
Ingvardsen CR, Schejbel B, Lübberstedt T (2008) Functional markers for resistance breeding. In: Lüttge U, Beyschlag W, Murata J (eds) Progress in botany, vol 69, part 2. Springer, Berlin, pp 61–87
Jacobs MMJ, van den Berg RG, Vleeshouwers VGAA, Visser M, Mank R, Sengers M, Hoekstra R, Vosman B (2008) AFLP analysis reveals a lack of phylogenetic structure within Solanum section. Petota BMC Evol Biol 8:1–12
Kuhl JC, Hanneman RE, Havey MJ (2001) Characterization and mapping of Rpi1, a late-blight resistance locus from diploid (1EBN) Mexican Solanum pinnatisectum. Mol Genet Genomics 265:977–985
Liu Z, Halterman D (2006) Identification and characterization of RB-orthologous genes from the late blight resistant wild potato species Solanum verrucosum. Physiol Mol Plant Pathol 69:230–239
Mantovani P, Van der Linden G, Maccaferri M, Danguineti MC, Tuberosa R (2006) Nucleotide-binding site (NBS) profiling of genetic diversity in durum wheat. Genome 49:1473–1480
McGuire S (2009) S. lycopersicum DNA sequence from clone LE_HBa-132O11 on chromosome 4, complete sequence. NCBI
Meyers BC, Dickerman AW, Michelmore RW, Sivaramakrisham S, Sobral BW, Young ND (1999) Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J 20:317–332
Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW (2003) Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15:809–834
Michelmore RW, Meyers BC (1998) Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res 8:1113–1130
Michelmore RW, Paran I, Kesseli RV (1991) Identification of markers linked to disease resistance genes by bulked segregant analysis. A rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci 88:9828–9832
Monosi B, Wisser R, Pennill L, Hulbert S (2004) Full-genome analysis of resistance gene homologues in rice. Theor Appl Genet 109:1434–1447
Mueller LA, Tanksley SD, Giovannoni JJ, van Eck J, Stack S, Choi D, Dong Kim B, Chen M, Cheng Z, Li C, Ling H, Xue Y, Seymour G, Bishop G, Bryan G, Sharma R, Khurana J, Tyagi A, Chattopadhyay D, Singh NK, Stiekema W, Lindhout P, Jesse T, Klein Lankhorst R, Bouzayen M, Shibata D, Tabata S, Granell A, Botella MA, Giuliano G, Frusciante L, Causse M, Zamir D (2005) The tomato sequencing project, the first cornerstone of the International Solanaceae Project (SOL). Comp Funct Genomics 6:153–158
Mueller LA, Giovannoni JJ, van Eck J, Stack S, Tanksley SD (2008) Solanum lycopersicum chromosome 11 clone C11HBa0119D16, complete sequence, ACCESSION AC171735, tomato genome sequencing. In: Submitted (12-MAR-2008) Plant Breed Gen, Cornell University, Emerson Hall, Ithaca, NY 14853, USA. NCBI
Naess SK, Bradeen JM, Wielgus SM, Haberlach GT, McGrath JM, Helgeson JP (2000) Resistance to late blight in Solanum bulbocastanum is mapped to chromosome 8. Theor Appl Genet 101:697–704
Park TH, Gros J, Sikkema A, Vleeshouwers VGAA, Muskens M, Allefs S, Jacobsen E, Visser RGF, van der Vossen EAG (2005a) The late blight resistance locus Rpi-blb3 from Solanum bulbocastanum belongs to a major late blight R gene cluster on chromosome 4 of potato. Mol Plant Microbe Interact 18:722–729
Park TH, Vleeshouwers V, Hutten RCB, van Eck HJ, van der Vossen E, Jacobsen E, Visser RGF (2005b) High-resolution mapping and analysis of the resistance locus Rpi-abpt against Phytophthora infestans in potato. Mol Breed 16:33–43
Pel MA, Foster SJ, Park T-H, Rietman H, Van Arkel G, Jones JDG, Van Eck HJ, Jacobsen E, Visser RGF, Van der Vossen EAG (2009) Mapping and Cloning of late blight resistance genes from Solanum venturii using an interspecific candidate gene approach. Mol Plant Microbe Interact 22:601–615
Pflieger S, Lefebvre V, Caranta C, Blattes A, Goffinet B, Palloix A (1999) Disease resistance gene analogs as candidates for QTLs involved in pepper-pathogen interactions. Genome 42:1100–1110
Rauscher GM, Smart CD, Simko I, Bonierbale M, Mayton H, Greenland A, Fry WE (2006) Characterization and mapping of Rpi-ber, a novel potato late blight resistance gene from Solanum berthaultii. Theor Appl Genet 112:674–687
Rivera-Peña A (1990) Wild tuber-bearing species of Solanum and incidence of Phytophthora infestans (Mont.) de Bary on the Western slopes of the volcano Nevado de Toluca. 2. Distribution of Phytophthora infestans. Potato Res 33:341–347
Rivera-Peña A, Molina-Galan J (1989) Wild tuber-bearing species of Solanum and incidence of Phytophthora infestans (Mont.) de Bary on the western slopes of the volcano Nevado de Toluca. 1. Solanum species. Potato Res 32:181–195
Ruiz de Galarreta JI, Carrasco A, Salazar A, Barrena I, Iturritxa E, Marquinez R, Legorburu FJ, Ritter E (1998) Wild Solanum species as resistance sources against different pathogens of potato. Potato Res 41:57–68
Sandbrink JM, Colon LT, Wolters PJCC, Stiekema WJ (2000) Two related genotypes of Solanum microdontum carry different segregating alleles for field resistance to Phytophthora infestans. Mol Breed 6:215–225
Seah S, Tellen AC, Williamson VM (2007) Introgressed and endogenous Mi-1 gene clusters in tomato differ by complex rearrangements in flanking sequences and show sequence exchange and diversifying selection among homologues. Theor Appl Genet 114:1289–1302
Śliwka J, Jakuczun H, Lebecka R, Marczewski W, Gebhardt C, Zimnoch-Guzowska E (2006) The novel, major locus Rpi-phu1 for late blight resistance maps to potato chromosome IX and is not correlated with long vegetation period. Theor Appl Genet 113:685–695
Smilde WD, Brigneti G, Jagger L, Perkins S, Jones JDG (2005) Solanum mochiquense chromosome IX carries a novel late blight resistance gene Rpi-moc1. Theor Appl Genet 110:252–258
Song J, Bradeen JM, Naess SK, Raasch JA, Wielgus SM, Haberlach GT, Liu J, Kuang H, Austin-Phillips S, Buell CR, Helgeson JP, Jiang J (2003) Gene RB cloned from Solanum bulbocastanum confers broad spectrum resistance to potato late blight. Proc Natl Acad Sci 100:9128–9133
Tan MYA, Hutten RCB, Celis C, Park TH, Niks RE, Visser RGF, van Eck HJ (2008) The RPi-mcd1 locus from Solanum microdontum involved in resistance to Phytophthora infestans, causing a delay in infection, maps on potato chromosome 4 in a cluster of NBS-LRR genes. Mol Plant Microbe Interact 21:909–918
Tanksley SD, Ganal MW, Prince JP, de-Vicente MC, Bonierbale MW, Broun P, Fulton TM, Giovannoni JJ, Grandillo S, Martin GB, Messeguer R, Miller JC, Miller L, Paterson AH, Pineda O, Roder MS, Wing RA, Wu W, Young ND (1992) High density molecular linkage maps of the tomato and potato genomes. Genetics 132:1141–1160
van der Linden CG, Wouters D, Mihalka V, Kochieva EZ, Smulders MJM, Vosman B (2004) Efficient targeting of plant disease resistance loci using NBS profiling. Theor Appl Genet 109:384–393
van der Vossen E, Sikkema A, Hekkert BTL, Gros J, Stevens P, Muskens M, Wouters D, Pereira A, Stiekema W, Allefs S (2003) An ancient R gene from the wild potato species Solanum bulbocastanum confers broad-spectrum resistance to Phytophthora infestans in cultivated potato and tomato. Plant J 36:867–882
van der Vossen EAG, Gros J, Sikkema A, Muskens M, Wouters D, Wolters P, Pereira A, Allefs S (2005) The Rpi-blb2 gene from Solanum bulbocastanum is an Mi-1 gene homolog conferring broad-spectrum late blight resistance in potato. Plant J 44:208–222
Van Soest L, Schöber B, Tazelaar M (1984) Resistance to Phytophthora infestans in tuber-bearing species of Solanum and its geographical distribution. Potato Res 27:393–411
Villamon FG, Spooner DM, Orrillo M, Mihovilovich E, Pérez W, Bonierbale M (2005) Late blight resistance linkages in a novel cross of the wild potato species Solanum paucissectum (series Piurana). Theor Appl Genet 111:1201–1214
Vleeshouwers VGAA, van Dooijeweert W, Keizer LCP, Sijpkes L, Govers F, Colon LT (1999) A laboratory assay for Phytophthora infestans resistance in various Solanum species reflects the field situation. Eur J Plant Pathol 105:241–250
Vleeshouwers VGAA, Rietman H, Krenek P, Champouret N, Young C, Oh S-K, Wang M, Bouwmeester K, Vosman B, Visser RGF, Jacobsen E, Govers F, Kamoun S, van der Vossen EAG (2008) Effector genomics accelerates discovery and functional profiling of potato disease resistance and Phytophthora infestans avirulence Genes. PLoS ONE 3(8):e2875. doi:10.1371/journal.pone.0002875
Wang M, Allefs S, van den Berg R, Vleeshouwers VGAA, van der Vossen EAG, Vosman B (2008a) Allele mining in Solanum : conserved homologues of Rpi-blb1 are identified in Solanum stoloniferum. Theor Appl Genet 116:933–943
Wang M, van den Berg RG, van der Linden GC, Vosman B (2008b) The utility of NBS profiling for plant systematics: a first study in tuber-bearing Solanum species. Plant Syst Evol 276:137–148
Wastie RL (1991) Breeding for resistance. In: Ingram DS, Williams PH (eds) Phytophthora infestans, the cause of late blight of potato. Advances in plant pathology, vol 7. Academic Press Ltd, London, pp 193–224
Zhang JF, Yuan YL, Niu C, Hinchliffe DJ, Lu Y, Yu S, Percy RG, Ulloa M, Cantrell RG (2007) AFLP-RGA markers in comparison with RGA and AFLP in cultivated tetraploid cotton. Crop Sci 47:180–187
Acknowledgments
This project was co-financed by the research program of the Centre of Biosystems Genomics (CBSG) which is part of the Netherlands Genomics Initiative/Netherlands Organization for Scientific Research. We thank Agrico Research B.V. for kindly providing populations ver03-292 and ver03-394. Furthermore, we thank the following persons for their contribution to the experimental part of this project: Roel Hoekstra (Centre for Genetic Resources, The Netherlands), Gerard van der Linden, Linda Kodde, Martijn van Kaauwen, Christel Denneboom, Dirk Budding, Ronald Hutten (all WUR Plant Breeding, Wageningen University and Research Centre), Guus Heselmans (Meijer B.V. The Netherlands) and Sjefke Allefs (Agrico Research B.V.). Gerard van der Linden and Sjefke Allefs are thanked for useful comments on earlier versions of the manuscript.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by C. Gebhardt.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Jacobs, M.M.J., Vosman, B., Vleeshouwers, V.G.A.A. et al. A novel approach to locate Phytophthora infestans resistance genes on the potato genetic map. Theor Appl Genet 120, 785–796 (2010). https://doi.org/10.1007/s00122-009-1199-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-009-1199-7