Identifying and Quantifying the Abundance of Economically Important Palms in Tropical Moist Forest Using UAV Imagery
Abstract
:1. Introduction
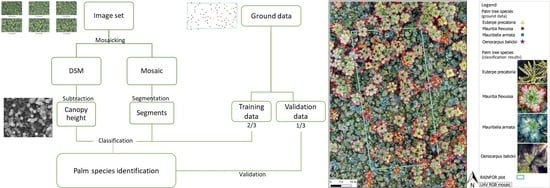
2. Materials and Methods
2.1. Study Area
2.2. Data Collection
2.3. Data Processing
3. Results
3.1. Palm Species Identification
3.2. Feature Selection
3.3. Palm Tree Species Quantification
4. Discussion
4.1. Palm Tree Identification and Classification Results
4.2. Feature Selection
4.3. Palm Tree Quantification and Validation Data
4.4. Considerations for Image Acquisition
4.5. Further Implications
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Plot | Mission | Flying Height AGL (m) | Flying Height ACL (m) | Area Covered (ha) | No. Total Images | Acquisition Date | Cloud Cover | Solar Elevation (°) | Wind Speed |
|---|---|---|---|---|---|---|---|---|---|
| JEN-14 | JEN-14_1 | 90 | 70 | 1.00 | 24 | 18-10-17 | overcast | 31.73 | calm |
| JEN-14 | JEN-14_2 | 50 | 30 | 1.05 | 66 | 18-10-17 | partly cloudy | 35.17 | calm |
| JEN-14 | JEN-14_3 | 90 | 70 | 1.67 | 19 | 18-10-17 | overcast | 54.12 | calm |
| JEN-14 | JEN-14_4 | 90 | 70 | 1.05 | 24 | 15-12-17 | partly cloudy | 56.97 | calm |
| JEN-14 | JEN-14_5 | 65 | 45 | 1.33 | 60 | 15-12-17 | partly cloudy | 57.76 | > 3 m/s |
| PIU-02 | PIU-02_1 | 90 | 70 | 3.63 | 95 | 26-11-17 | clear sky | 49.59 | calm |
| PIU-02 | PIU-02_2 | 65 | 45 | 3.22 | 95 | 26-11-17 | clear sky | 54.57 | calm |
| PRN-01 | PRN-01_1 | 90 | 70 | 3.84 | 86 | 20-11-17 | clear sky | 59.70 | medium |
| PRN-01 | PRN-01_2 | 60 | 40 | 2.03 | 92 | 20-11-17 | partly cloudy | 65.16 | calm |
| QUI-01 | QUI-01_1 | 90 | 70 | 3.35 | 94 | 09-12-17 | partly cloudy | 56.84 | calm |
| QUI-01 | QUI-01_2 | 65 | 45 | 2.60 | 85 | 09-12-17 | clear sky | 66.36 | calm |
| SAM-01 | SAM-01_1 | 90 | 70 | 1.23 | 35 | 18-11-17 | clear sky | 51.20 | calm |
| SAM-01 | SAM-01_2 | 90 | 70 | 1.12 | 30 | 18-11-17 | clear sky | 55.88 | calm |
| SAM-01 | SAM-01_3 | 60 | 40 | 1.12 | 61 | 18-11-17 | clear sky | 56.97 | calm |
| VEN-01 | VEN-01_1 | 90 | 70 | 0.84 | 27 | 06-10-17 | partly cloudy | 85.39 | calm |
| VEN-01 | VEN-01_2 | 65 | 45 | 0.98 | 50 | 06-10-17 | partly cloudy | 86.86 | calm |
| VEN-02 | VEN-02_1 | 90 | 70 | 0.69 | 47 | 05-10-17 | clear sky | 29.87 | calm |
| VEN-02 | VEN-02_2 | 60 | 40 | 0.69 | 84 | 05-10-17 | clear sky | 27.88 | calm |
| VEN-02 | VEN-02_4 | 65 | 45 | 1.76 | 46 | 06-10-17 | clear sky | 40.76 | calm |
| VEN-03 | VEN-03_2 | 90 | 70 | 0.79 | 47 | 06-10-17 | partly cloudy | 52.56 | calm |
| VEN-03 | VEN-03_3 | 65 | 45 | 0.79 | 79 | 06-10-17 | partly cloudy | 55.30 | calm |
| VEN-04 | VEN-04_1 | 90 | 70 | 0.91 | 46 | 05-10-17 | clear sky | 81.38 | calm |
| VEN-04 | VEN-04_2 | 65 | 45 | 0.81 | 69 | 06-10-17 | partly cloudy | 41.86 | calm |
| VEN-05 | VEN-05_1 | 90 | 70 | 1.29 | 64 | 05-10-17 | partly cloudy | 46.76 | calm |
| VEN-05 | VEN-05_2 | 65 | 45 | 0.93 | 83 | 05-10-17 | partly cloudy | 53.23 | calm |
Appendix B


Appendix C
| Plot | k-NN | RP | RF | SVMR | ||||
|---|---|---|---|---|---|---|---|---|
| Acc. | Κ | Acc. | κ | Acc. | Κ | Acc. | κ | |
| JEN-14 | 0.86 | 0.83 | 0.82 | 0.78 | 0.90 | 0.88 | 0.89 | 0.87 |
| PIU-02 | 0.82 | 0.78 | 0.82 | 0.77 | 0.89 | 0.86 | 0.89 | 0.86 |
| PRN-01 | 0.59 | 0.54 | 0.65 | 0.60 | 0.85 | 0.83 | 0.89 | 0.88 |
| QUI-01 | 0.64 | 0.53 | 0.69 | 0.59 | 0.79 | 0.72 | 0.72 | 0.63 |
| SAM-01 | 0.71 | 0.65 | 0.71 | 0.65 | 0.86 | 0.83 | 0.89 | 0.87 |
| VEN-01 | 0.61 | 0.54 | 0.68 | 0.62 | 0.84 | 0.81 | 0.85 | 0.82 |
| VEN-02 | 0.69 | 0.64 | 0.72 | 0.67 | 0.83 | 0.80 | 0.87 | 0.85 |
| VEN-03 | 0.72 | 0.65 | 0.75 | 0.69 | 0.86 | 0.83 | 0.79 | 0.74 |
| VEN-04 | 0.65 | 0.53 | 0.72 | 0.63 | 0.81 | 0.75 | 0.78 | 0.71 |
| VEN-05 | 0.68 | 0.57 | 0.82 | 0.76 | 0.88 | 0.84 | 0.89 | 0.86 |
| Mean | 0.70 | 0.62 | 0.74 | 0.68 | 0.85 | 0.82 | 0.85 | 0.81 |
| Plot | k-NN | RP | RF | SVMR | ||||
|---|---|---|---|---|---|---|---|---|
| Acc. | κ | Acc. | κ | Acc. | κ | Acc. | κ | |
| JEN-14 | 0.69 | 0.61 | 0.83 | 0.79 | 0.85 | 0.81 | 0.85 | 0.83 |
| PIU-02 | 0.65 | 0.57 | 0.76 | 0.70 | 0.77 | 0.71 | 0.77 | 0.71 |
| PRN-01 | 0.55 | 0.50 | 0.58 | 0.53 | 0.78 | 0.75 | 0.78 | 0.75 |
| QUI-01 | 0.55 | 0.41 | 0.70 | 0.60 | 0.73 | 0.64 | 0.73 | 0.64 |
| SAM-01 | 0.61 | 0.52 | 0.68 | 0.60 | 0.88 | 0.85 | 0.82 | 0.77 |
| VEN-01 | 0.50 | 0.40 | 0.67 | 0.61 | 0.77 | 0.72 | 0.77 | 0.72 |
| VEN-02 | 0.53 | 0.45 | 0.66 | 0.60 | 0.71 | 0.66 | 0.71 | 0.66 |
| VEN-03 | 0.63 | 0.53 | 0.74 | 0.67 | 0.79 | 0.72 | 0.79 | 0.74 |
| VEN-04 | 0.61 | 0.47 | 0.77 | 0.70 | 0.77 | 0.69 | 0.77 | 0.69 |
| VEN-05 | 0.57 | 0.42 | 0.75 | 0.67 | 0.78 | 0.70 | 0.78 | 0.70 |
| Mean | 0.59 | 0.49 | 0.71 | 0.65 | 0.78 | 0.73 | 0.78 | 0.72 |
Appendix D
| Evaluation index | JEN14 | PIU02 | PRN01 | QUI01 | SAM01 | VEN01 | VEN02 | VEN03 | VEN04 | VEN05 |
|---|---|---|---|---|---|---|---|---|---|---|
| Number of correctly detected palm trees | 95 | 58 | 103 | 96 | 56 | 45 | 112 | 97 | 66 | 78 |
| Number of all the detected objects in the mosaic | 152 | 92 | 151 | 143 | 179 | 134 | 144 | 111 | 125 | 111 |
| Number of all the visible palm trees in the mosaic | 96 | 60 | 134 | 119 | 106 | 77 | 154 | 148 | 171 | 105 |
| Number of all the palm trees with a DBH higher than 10 cm (Ground data) | 128 | 76 | 197 | 204 | 123 | 132 | 268 | 196 | 221 | 123 |
| Precision (%) | 62.50 | 63.04 | 68.21 | 67.13 | 31.28 | 33.58 | 77.78 | 87.39 | 52.80 | 70.27 |
| Recall with Visible palms in the mosaic (%) | 98.96 | 96.67 | 76.87 | 80.67 | 52.83 | 58.44 | 72.73 | 65.54 | 38.60 | 74.29 |
| Recall with Ground data (%) | 74.22 | 76.32 | 52.28 | 47.06 | 45.53 | 34.09 | 41.79 | 49.49 | 29.86 | 63.41 |
| F1 score with Visible palms in the mosaic | 0.77 | 0.76 | 0.72 | 0.73 | 0.39 | 0.43 | 0.75 | 0.75 | 0.45 | 0.72 |
| F1 score with Ground data | 0.68 | 0.69 | 0.59 | 0.55 | 0.37 | 0.34 | 0.54 | 0.63 | 0.38 | 0.67 |
Appendix E
| Species/Plot | JEN14 | PIU02 | PRN01 | QUI01 | SAM01 | VEN01 | VEN02 | VEN03 | VEN04 | VEN05 | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|
| A. murumuru | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 |
| A. butyracea | 0 | 0 | 0 | 0 | 15 | 0 | 0 | 0 | 0 | 0 | 15 |
| E. indet | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| E. precatoria | 3 | 5 | 31 | 1 | 1 | 38 | 37 | 5 | 0 | 0 | 121 |
| Indet indet | 0 | 5 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7 |
| M. flexuosa | 124 | 71 | 109 | 88 | 104 | 71 | 184 | 180 | 129 | 80 | 1140 |
| M. armata | 0 | 0 | 14 | 115 | 0 | 0 | 3 | 11 | 92 | 43 | 278 |
| Oenocarpus spp. | 0 | 0 | 6 | 0 | 0 | 1 | 2 | 0 | 0 | 0 | 9 |
| S. exorrhiza | 1 | 0 | 34 | 0 | 3 | 22 | 42 | 0 | 0 | 0 | 102 |
| Total | 128 | 82 | 199 | 204 | 123 | 132 | 268 | 196 | 221 | 123 | 1676 |
Appendix F
| Mission | Mosaic Area (ha) | Segmentation | Texture Extraction | Training Set | Classification | Quantification |
|---|---|---|---|---|---|---|
| JEN-14 | 0.77 | 3 min | 27 min | 6 min | 12 min | 4 min |
| PIU-02 | 2.14 | 6 min | 50 min | 20 min | 24 min | 5 min |
| PRN-01 | 2.15 | 4 min | 32 min | 20 min | 18 min | 5 min |
| QUI-01 | 3.13 | 13 min | 30 min | 10 min | 16 min | 6 min |
| SAM-01 | 0.99 | 5 min | 26 min | 47 min | 16 min | 1 min |
| VEN-01 | 1.45 | 4 min | 39 min | 10 min | 24 min | 1 min |
| VEN-02 | 1.32 | 7 min | 43 min | 1 h 20 min | 19 min | 5 min |
| VEN-03 | 1.35 | 9 min | 25 min | 6 min | 13 min | 12 min |
| VEN-04 | 1.04 | 3 min | 21 min | 43 min | 3 h 42 min | 1 min |
| VEN-05 | 2.31 | 10 min | 34 min | 12 min | 14 min | 8 min |
References
- Eiserhardt, W.L.; Svenning, J.C.; Kissling, W.D.; Balslev, H. Geographical ecology of the palms (Arecaceae): Determinants of diversity and distributions across spatial scales. Ann. Bot. 2011, 108, 1391–1416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Couvreur, T.L.P.; Baker, W.J. Tropical rain forest evolution: Palms as a model group. BMC Biol. 2013, 11, 2–5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, N. Palms and People in the Amazon; Geobotany Studies; Springer International Publishing: Cham, Switzerland, 2015; ISBN 978-3-319-05508-4. [Google Scholar]
- Vormisto, J. Palms as rainforest resources: How evenly are they distributed in Peruvian Amazonia? Biodivers. Conserv. 2002, 11, 1025–1045. [Google Scholar] [CrossRef]
- Horn, C.M.; Vargas Paredes, V.H.; Gilmore, M.P.; Endress, B.A. Spatio-temporal patterns of Mauritia flexuosa fruit extraction in the Peruvian Amazon: Implications for conservation and sustainability. Appl. Geogr. 2018, 97, 98–108. [Google Scholar] [CrossRef]
- Roucoux, K.H.; Lawson, I.T.; Baker, T.R.; Del Castillo Torres, D.; Draper, F.C.; Lähteenoja, O.; Gilmore, M.P.; Honorio Coronado, E.N.; Kelly, T.J.; Mitchard, E.T.A.; et al. Threats to intact tropical peatlands and opportunities for their conservation. Conserv. Biol. 2017, 31, 1283–1292. [Google Scholar] [CrossRef] [Green Version]
- Draper, F.C.; Roucoux, K.H.; Lawson, I.T.; Mitchard, E.T.A.; Honorio Coronado, E.N.; Lähteenoja, O.; Torres Montenegro, L.; Valderrama Sandoval, E.; Zaráte, R.; Baker, T.R. The distribution and amount of carbon in the largest peatland complex in Amazonia. Environ. Res. Lett. 2014, 9, 124017. [Google Scholar] [CrossRef]
- Virapongse, A.; Endress, B.A.; Gilmore, M.P.; Horn, C.; Romulo, C. Ecology, livelihoods, and management of the Mauritia flexuosa palm in South America. Glob. Ecol. Conserv. 2017, 10, 70–92. [Google Scholar] [CrossRef]
- Nobre, C.A.; Sampaio, G.; Borma, L.S.; Castilla-Rubio, J.C.; Silva, J.S.; Cardoso, M. Land-use and climate change risks in the Amazon and the need of a novel sustainable development paradigm. Proc. Natl. Acad. Sci. USA 2016, 113, 10759–10768. [Google Scholar] [CrossRef] [Green Version]
- Monitoring of the Andean Amazon Project (MAAP). Deforestation Hotspots in the Peruvian Amazon, 2012–2014|MAAP—Monitoring of the Andean Amazon Project. Available online: http://maaproject.org/2018/hotspots-peru2017/ (accessed on 21 February 2018).
- Falen Horna, L.Y.; Honorio Coronado, E.N. Evaluación de las técnicas de aprovechamiento de frutos de aguaje (Mauritia flexuosa L.f.) en el distrito de Jenaro Herrera, Loreto, Perú. Folia Amaz. 2019, 27, 131–150. [Google Scholar] [CrossRef]
- Nevalainen, O.; Honkavaara, E.; Tuominen, S.; Viljanen, N.; Hakala, T.; Yu, X.; Hyyppä, J.; Saari, H.; Pölönen, I.; Imai, N.N.; et al. Individual tree detection and classification with UAV-Based photogrammetric point clouds and hyperspectral imaging. Remote Sens. 2017, 9, 185. [Google Scholar] [CrossRef] [Green Version]
- IIAP. Diversidad de Vegetación de la Amazonía Peruana Expresada en un Mosaico de Imágenes de Satélite; BIODAMAZ: Iquitos, Peru, 2004. [Google Scholar]
- Lhteenoja, O.; Page, S. High diversity of tropical peatland ecosystem types in the Pastaza-Maraón basin, Peruvian Amazonia. J. Geophys. Res. Biogeosci. 2011, 116, G02025. [Google Scholar]
- Fuyi, T.; Boon Chun, B.; Mat Jafri, M.Z.; Hwee San, L.; Abdullah, K.; Mohammad Tahrin, N. Land cover/use mapping using multi-band imageries captured by Cropcam Unmanned Aerial Vehicle Autopilot (UAV) over Penang Island, Malaysia. In Proceedings of the SPIE; Carapezza, E.M., White, H.J., Eds.; SPIE: Edinburgh, UK, 2012; Volume 8540, p. 85400S. [Google Scholar]
- Petrou, Z.I.; Stathaki, T. Remote sensing for biodiversity monitoring: A review of methods for biodiversity indicator extraction and assessment of progress towards international targets. Biodivers. Conserv. 2015, 24, 2333–2363. [Google Scholar] [CrossRef]
- Rocchini, D.; Boyd, D.S.; Féret, J.-B.; Foody, G.M.; He, K.S.; Lausch, A.; Nagendra, H.; Wegmann, M.; Pettorelli, N. Satellite remote sensing to monitor species diversity: Potential and pitfalls. Remote Sens. Ecol. Conserv. 2016, 2, 25–36. [Google Scholar] [CrossRef]
- Liu, Z.; Zhang, Y.; Yu, X.; Yuan, C. Unmanned surface vehicles: An overview of developments and challenges. Annu. Rev. Control 2016, 41, 71–93. [Google Scholar] [CrossRef]
- Matese, A.; Toscano, P.; Di Gennaro, S.; Genesio, L.; Vaccari, F.; Primicerio, J.; Belli, C.; Zaldei, A.; Bianconi, R.; Gioli, B. Intercomparison of UAV, Aircraft and Satellite Remote Sensing Platforms for Precision Viticulture. Remote Sens. 2015, 7, 2971–2990. [Google Scholar] [CrossRef] [Green Version]
- Cruzan, M.B.; Weinstein, B.G.; Grasty, M.R.; Kohrn, B.F.; Hendrickson, E.C.; Arredondo, T.M.; Thompson, P.G. Small Unmanned Aerial Vehicles (Micro-Uavs, Drones) in Plant Ecology. Appl. Plant Sci. 2016, 4, 1600041. [Google Scholar] [CrossRef]
- Salamí, E.; Barrado, C.; Pastor, E. UAV Flight Experiments Applied to the Remote Sensing of Vegetated Areas. Remote Sens. 2014, 6, 11051–11081. [Google Scholar] [CrossRef] [Green Version]
- Calderón, R.; Navas-Cortés, J.A.; Lucena, C.; Zarco-Tejada, P.J. High-resolution airborne hyperspectral and thermal imagery for early detection of Verticillium wilt of olive using fluorescence, temperature and narrow-band spectral indices. Remote Sens. Environ. 2013, 139, 231–245. [Google Scholar] [CrossRef]
- Zarco-Tejada, P.J.; Morales, A.; Testi, L.; Villalobos, F.J. Spatio-temporal patterns of chlorophyll fluorescence and physiological and structural indices acquired from hyperspectral imagery as compared with carbon fluxes measured with eddy covariance. Remote Sens. Environ. 2013, 133, 102–115. [Google Scholar] [CrossRef]
- Laliberte, A.S.; Goforth, M.A.; Steele, C.M.; Rango, A. Multispectral Remote Sensing from Unmanned Aircraft: Image Processing Workflows and Applications for Rangeland Environments. Remote Sens. 2011, 3, 2529–2551. [Google Scholar] [CrossRef] [Green Version]
- Inc., F.S.; Puliti, S.; Ørka, H.O.; Gobakken, T.; Næsset, E.; Nevalainen, O.; Honkavaara, E.; Tuominen, S.; Viljanen, N.; Hakala, T.; et al. Use of a multispectral UAV photogrammetry for detection and tracking of forest disturbance dynamics. Remote Sens. 2015, 7, 37–46. [Google Scholar]
- Iglhaut, J.; Cabo, C.; Puliti, S.; Piermattei, L.; O’Connor, J.; Rosette, J. Structure from Motion Photogrammetry in Forestry: A Review. Curr. For. Rep. 2019, 5, 155–168. [Google Scholar] [CrossRef] [Green Version]
- Lou, X.; Huang, D.; Fan, L.; Xu, A. An Image Classification Algorithm Based on Bag of Visual Words and Multi-kernel Learning. J. Multimed. 2014, 9, 269–277. [Google Scholar] [CrossRef] [Green Version]
- Csillik, O.; Cherbini, J.; Johnson, R.; Lyons, A.; Kelly, M. Identification of Citrus Trees from Unmanned Aerial Vehicle Imagery Using Convolutional Neural Networks. Drones 2018, 2, 39. [Google Scholar] [CrossRef] [Green Version]
- Blaschke, T. Object based image analysis for remote sensing. ISPRS J. Photogramm. Remote Sens. 2010, 65, 2–16. [Google Scholar] [CrossRef] [Green Version]
- Feng, Q.; Liu, J.; Gong, J. UAV Remote Sensing for Urban Vegetation Mapping Using Random Forest and Texture Analysis. Remote Sens. 2015, 7, 1074–1094. [Google Scholar] [CrossRef] [Green Version]
- Momsen, E.; Metz, M. GRASS GIS Manual: I. Segment. Available online: https://grass.osgeo.org/grass74/manuals/i.segment.html (accessed on 28 February 2018).
- Baena, S.; Moat, J.; Whaley, O.; Boyd, D.S. Identifying species from the air: UAVs and the very high resolution challenge for plant conservation. PLoS ONE 2017, 12, e0188714. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Ribera, J.; Boomsma, C.; Delp, E.J. Plant leaf segmentation for estimating phenotypic traits. In Proceedings of the International Conference on Image Processing (ICIP), Beijing, China, 17–20 September 2018; Volume 2017, pp. 3884–3888. [Google Scholar]
- Blaschke, T.; Hay, G.J.; Kelly, M.; Lang, S.; Hofmann, P.; Addink, E.; Queiroz Feitosa, R.; van der Meer, F.; van der Werff, H.; van Coillie, F.; et al. Geographic Object-Based Image Analysis—Towards a new paradigm. ISPRS J. Photogramm. Remote Sens. 2014, 87, 180–191. [Google Scholar] [CrossRef] [Green Version]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I. Textural Features for Image Classification. IEEE Trans. Syst. Man. Cybern. 1973, SMC-3, 610–621. [Google Scholar] [CrossRef] [Green Version]
- Mellor, A.; Haywood, A.; Stone, C.; Jones, S. The Performance of Random Forests in an Operational Setting for Large Area Sclerophyll Forest Classification. Remote Sens. 2013, 5, 2838–2856. [Google Scholar] [CrossRef] [Green Version]
- Fassnacht, F.E.; Latifi, H.; Stereńczak, K.; Modzelewska, A.; Lefsky, M.; Waser, L.T.; Straub, C.; Ghosh, A. Review of studies on tree species classification from remotely sensed data. Remote Sens. Environ. 2016, 186, 64–87. [Google Scholar] [CrossRef]
- Lu, D.; Weng, Q. A survey of image classification methods and techniques for improving classification performance. Int. J. Remote Sens. 2007, 28, 823–870. [Google Scholar] [CrossRef]
- Meng, Q.; Cieszewski, C.J.; Madden, M.; Borders, B.E. K Nearest Neighbor Method for Forest Inventory Using Remote Sensing Data. GIScience Remote Sens. 2007, 44, 149–165. [Google Scholar] [CrossRef]
- Pennington, T.D.; Reynel, C.; Daza, A.; Wise, R. Illustrated Guide to the Trees of Peru; Hunt, D., Ed.; University of Michigan: Ann Arbor, MI, USA, 2004; ISBN 0953813436. [Google Scholar]
- Henderson, A.; Galeano, G.; Bernal, R. Field Guide to the Palms of the Americas; Princeton University Press: Princeton, NJ, USA, 1995; ISBN 978-0-6916-5612-0. [Google Scholar]
- Ter Steege, H.; Pitman, N.C.; Sabatier, D.; Baraloto, C.; Salomão, R.P.; Guevara, J.E.; Phillips, O.L.; Castilho, C.V.; Magnusson, W.E.; Molino, J.-F.; et al. Hyperdominance in the Amazonian tree flora. Science 2013, 342, 1243092. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Da Silveira Agostini-Costa, T. Bioactive compounds and health benefits of some palm species traditionally used in Africa and the Americas—A review. J. Ethnopharmacol. 2018, 224, 202–229. [Google Scholar] [CrossRef] [PubMed]
- Malhi, Y.; Phillips, O.L.; Lloyd, J.; Baker, T.; Wright, J.; Almeida, S.; Arroyo, L.; Frederiksen, T.; Grace, J.; Higuchi, N.; et al. An international network to monitor the structure, composition and dynamics of Amazonian forests (RAINFOR). J. Veg. Sci. 2002, 13, 439–450. [Google Scholar] [CrossRef]
- Lopez-Gonzalez, G.; Lewis, S.L.; Burkitt, M.; Phillips, O.L.; Baker, T.R.; Phillips, O. ForestPlots.net Database. Available online: https://www.forestplots.net/secure/ (accessed on 8 November 2017).
- Lopez-Gonzalez, G.; Lewis, S.L.; Burkitt, M.; Phillips, O.L. ForestPlots.net: A web application and research tool to manage and analyse tropical forest plot data. J. Veg. Sci. 2011, 22, 610–613. [Google Scholar] [CrossRef]
- DJI. DJI Phantom 4 Pro—Photography Drone. Available online: https://www.dji.com/dk/phantom-4-pro/info%0A; https://www.dji.com/ae/phantom-4-pro (accessed on 12 February 2018).
- Pix4D. How to Improve the Outputs of Dense Vegetation Areas?—Support. Available online: https://support.pix4d.com/hc/en-us/articles/202560159-How-to-improve-the-outputs-of-dense-vegetation-areas- (accessed on 14 February 2018).
- R Core Development Team. R: A Language and Environment for Statistical Computing; R foundation for Statistical Computing: Vienna, Austria, 2013; Available online: http://www.R-project.org/ (accessed on 20 February 2018).
- Colomina, I.; Molina, P. Unmanned aerial systems for photogrammetry and remote sensing: A review. ISPRS J. Photogramm. Remote Sens. 2014, 92, 79–97. [Google Scholar] [CrossRef] [Green Version]
- Niederheiser, R.; MokroA, M.; Lange, J.; Petschko, H.; Prasicek, G.; Elberink, S.O. Deriving 3D point clouds from terrestrial photographs—Comparison of different sensors and software. In Proceedings of the International Archives of the Photogrammetry, Remote Sensing and Spatial Information Sciences—ISPRS Archives, Prague, Czech Republic, 12–19 July 2016; Volume 41, pp. 685–692. [Google Scholar]
- Bivand, R. Interface between GRASS 7 Geographical Information System and R [R Package Rgrass7 Version 0.1-12]. 2018. Available online: https://cran.r-project.org/web/packages/rgrass7/index.html (accessed on 24 April 2019).
- Hijmans, R.; Van Ettern, J.; Cheng, J.; Mattiuzzi, M.; Sumner, M.; Greenberg, J.; Perpinan, O.; Bevan, A.; Racine, E.; Shortridge, A.; et al. Package “Raster”: Geographic Data Analysis and Modeling; The R Foundation: Vienna, Austria, 2017. [Google Scholar]
- Kuhn, M. Building Predictive Models in R Using the caret Package. J. Stat. Softw. 2008, 28, 1–26. [Google Scholar] [CrossRef] [Green Version]
- Mallinis, G.; Koutsias, N.; Tsakiri-Strati, M.; Karteris, M. Object-based classification using Quickbird imagery for delineating forest vegetation polygons in a Mediterranean test site. ISPRS J. Photogramm. Remote Sens. 2008, 63, 237–250. [Google Scholar] [CrossRef]
- Pebesma, E. Simple Features for R: Standardized Support for Spatial Vector Data [R Package sf Version 0.8-0]. R J. 2018, 10, 439–446. [Google Scholar] [CrossRef] [Green Version]
- Duarte, L.; Silva, P.; Teodoro, A. Development of a QGIS Plugin to Obtain Parameters and Elements of Plantation Trees and Vineyards with Aerial Photographs. ISPRS Int. J. Geo-Inf. 2018, 7, 109. [Google Scholar] [CrossRef] [Green Version]
- Draper, F.C.; Baraloto, C.; Brodrick, P.G.; Phillips, O.L.; Martinez, R.V.; Honorio Coronado, E.N.; Baker, T.R.; Zárate Gómez, R.; Amasifuen Guerra, C.A.; Flores, M.; et al. Imaging spectroscopy predicts variable distance decay across contrasting Amazonian tree communities. J. Ecol. 2019, 107, 696–710. [Google Scholar] [CrossRef]
- Mulatu, K.; Mora, B.; Kooistra, L.; Herold, M. Biodiversity Monitoring in Changing Tropical Forests: A Review of Approaches and New Opportunities. Remote Sens. 2017, 9, 1059. [Google Scholar] [CrossRef] [Green Version]








| Plot | Max. Canopy Height (m) | Mean Canopy Height (m) | No. Stems | No. Palm Tree Stems | No. Palm Tree Species | Palm Tree Species | Dominant species | % M. flexuosa Abundance | Palm Visibility * (%) |
|---|---|---|---|---|---|---|---|---|---|
| JEN-14 | 34.8 | 18.7 | 234 | 128 | 3 | E. precatoria, S. exorrhiza, M. flexuosa | M. flexuosa | 53.0 | 75.0 |
| PIU-02 | 37.5 | 20.1 | 404 | 77 | 3 | E. precatoria, Elaeis sp., M. flexuosa | M. flexuosa | 17.3 | 77.9 |
| PRN-01 | 37.9 | 19.6 | 310 | 199 | 6 | E. precatoria, M. armata, A. murumuru, O. balickii, S. exorrhiza, M. flexuosa | M. flexuosa | 35.2 | 67.3 |
| QUI-01 | 29.1 | 15.75 | 398 | 204 | 3 | E. precatoria, M. armata, M. flexuosa | Tabebuia insignis | 22.1 | 58.3 |
| SAM-01 | 34.7 | 19 | 251 | 123 | 4 | E. precatoria, A. butyracea, S. exorrhiza, M. flexuosa | M. flexuosa | 41.0 | 86.2 |
| VEN-01 | 30.1 | 20.1 | 253 | 132 | 4 | E. precatoria, M. flexuosa, O. mapora, S. exorrhiza | M. flexuosa | 28.1 | 58.3 |
| VEN-02 | 30.1 | 16.7 | 326 | 268 | 5 | E. precatoria, M. flexuosa, M. armata, O. mapora, S. exorrhiza | M. flexuosa | 56.4 | 57.5 |
| VEN-03 | 30.1 | 15.38 | 254 | 196 | 3 | E. precatoria, M. armata, M. flexuosa | M. flexuosa | 70.9 | 75.5 |
| VEN-04 | 32.3 | 12.9 | 270 | 221 | 2 | M. flexuosa, M. armata | M. armata | 47.8 | 77.4 |
| VEN-05 | 28.1 | 16.6 | 248 | 124 | 2 | M. flexuosa, M. armata | Ilex andarensis | 32.7 | 84.7 |
| Plot | Flying Height AGL (m) | GSD (cm) | Area Covered (ha) | No. Images Used | 2D Keypoints (median per image) | Reproj. Error (pix) | Point Density (points/m2) | Point Density | Interpolation Method |
|---|---|---|---|---|---|---|---|---|---|
| JEN-14 | 90 | 1.41 | 1.84 | 71 | 75,496 | 0.231 | 5,421,087 | Optimal | IDW |
| PIU-02 | 90-65 | 1.9 | 5.36 | 191 | 71,505 | 0.180 | 84,420,906 | Optimal | IDW |
| PRN-01 | 90 | 1.87 | 3.58 | 76 | 77,853 | 0.265 | 34,899,971 | high/slow | IDW |
| QUI-01 | 90 | 2.09 | 5.09 | 94 | 75,794 | 0.239 | 2,729,608 | high/slow | IDW |
| SAM-01 | 90-60 | 1.84 | 1.73 | 40 | 74,923 | 0.218 | 13,797,799 | Optimal | IDW |
| VEN-01 | 90-65 | 1.28 | 1.96 | 73 | 74,312 | 0.216 | 7,362,904 | Optimal | Triangulation |
| VEN-02 | 90-60 | 1.22 | 2.48 | 188 | 75,201 | 0.245 | 34,667,002 | Optimal | IDW |
| VEN-03 | 90 | 2.06 | 9.27 | 168 | 74,250 | 0.218 | 5,292,017 | Optimal | IDW |
| VEN-04 | 65 | 1.62 | 1.84 | 69 | 78,824 | 0.207 | 120,548,930 | high/slow | IDW |
| VEN-05 | 90 | 2.06 | 3.49 | 60 | 76,969 | 0.205 | 31,389,942 | Optimal | IDW |
| Predictor | Description |
|---|---|
| Canopy height model | Height above the ground (meters) |
| Area | Area size of each segment |
| Compactness | Compactness of each segment, calculated as: |
| Fractal dimension | Fractal dimension of the boundary of each segment (Mandelbrot, 1982) |
| Mean RGB | Average of all the pixel values per segment per band |
| SD RGB | Standard deviation of all the pixel values per segment per band |
| Median RGB | Median of all the pixel values per segment per band |
| Max RGB | Maximum pixel value per segment per band |
| Min RGB | Minimum pixel value per segment per band |
| Mean of entropy RGB | Mean entropy values per segment (Haralick, 1979) |
| SD of entropy RGB | Standard deviation of the entropy values per segment (Haralick, 1979) |
| Mean of the Sum of Variance RGB | Mean of the sum of variance values per segment (Haralick, 1979) |
| SD of the Sum of Variance RGB | Standard deviation of the sum of variance values per segment (Haralick, 1979) |
| Reference | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Prediction | Trees | A. butyracea | E. precatoria | M. flexuosa | M. armata | A. murumuru | Oenocarpus spp. | S. exorrhiza | Water | Soil | Total |
| Trees | 497 | 5 | 2 | 72 | 25 | 0 | 0 | 5 | 4 | 4 | 614 |
| A. butyracea | 7 | 50 | 0 | 12 | 0 | 0 | 0 | 0 | 0 | 1 | 70 |
| E. precatoria | 13 | 0 | 369 | 7 | 2 | 1 | 3 | 10 | 0 | 1 | 406 |
| M. flexuosa | 69 | 8 | 13 | 489 | 20 | 1 | 1 | 26 | 1 | 3 | 631 |
| M. armata | 39 | 0 | 3 | 36 | 348 | 0 | 3 | 9 | 0 | 1 | 439 |
| A. murumuru | 2 | 0 | 2 | 2 | 0 | 62 | 0 | 1 | 0 | 0 | 69 |
| Oenocarpus spp. | 3 | 0 | 11 | 4 | 0 | 0 | 190 | 1 | 0 | 0 | 209 |
| S. exorrhiza | 19 | 2 | 6 | 32 | 1 | 2 | 1 | 216 | 0 | 0 | 279 |
| Water | 3 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 253 | 4 | 262 |
| Soil | 2 | 1 | 2 | 5 | 0 | 0 | 0 | 0 | 6 | 667 | 683 |
| Total | 654 | 67 | 408 | 660 | 396 | 66 | 198 | 268 | 264 | 681 | 2702 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tagle Casapia, X.; Falen, L.; Bartholomeus, H.; Cárdenas, R.; Flores, G.; Herold, M.; Honorio Coronado, E.N.; Baker, T.R. Identifying and Quantifying the Abundance of Economically Important Palms in Tropical Moist Forest Using UAV Imagery. Remote Sens. 2020, 12, 9. https://doi.org/10.3390/rs12010009
Tagle Casapia X, Falen L, Bartholomeus H, Cárdenas R, Flores G, Herold M, Honorio Coronado EN, Baker TR. Identifying and Quantifying the Abundance of Economically Important Palms in Tropical Moist Forest Using UAV Imagery. Remote Sensing. 2020; 12(1):9. https://doi.org/10.3390/rs12010009
Chicago/Turabian StyleTagle Casapia, Ximena, Lourdes Falen, Harm Bartholomeus, Rodolfo Cárdenas, Gerardo Flores, Martin Herold, Eurídice N. Honorio Coronado, and Timothy R. Baker. 2020. "Identifying and Quantifying the Abundance of Economically Important Palms in Tropical Moist Forest Using UAV Imagery" Remote Sensing 12, no. 1: 9. https://doi.org/10.3390/rs12010009