Abstract
To create more resilient growing systems for greenhouse crops, the efficacy of different organic amendments alone and in combination with the biocontrol strain Pseudomonas chlororaphis 4.4.1 was tested to suppress Pythium disease in tomato plants. Four independent greenhouse experiments were performed with young tomato plants in potting soil. Inoculating the pathogen Pythium aphanidermatum to the potting soil prior to sowing resulted in significant losses of tomato plants; i.e. 94–98% healthy plants in pathogen-free control compared to 43–68% healthy plants when the pathogen was added. P. chlororaphis 4.4.1 inoculations increased the number of healthy plants in the potting soil up to 80% on average; soil and seed treatment were both effective. Numbers of P. chlororaphis in the rhizosphere had increased 4 to 100 fold 3 weeks after its inoculation (qPCR detection). All compost types reduced Pythium disease in potting soil resulting in 80–95% healthy plants. Animal bone char was not effective against P. aphanidermatum, whereas with plant-based biochar there was an effect, although not significantly different from the control treatment. Phosphorous and potassium uptake by the plants were elevated by the different organic amendments. These results demonstrate the potential of the organic amendments to enhance sustainability of growing media such as potting soil by increasing its disease suppressiveness, the re-use of nutrients and replacement of peat by using organic amendments. In addition, inoculating the growing medium or tomato seeds with the biocontrol strain P. chlororaphis 4.4.1 enhanced Pythium control in the susceptible growing media.
Similar content being viewed by others
Introduction
With an increasing world population (predicted 9.7 × 109 in 2050 (United Nations 2017)) and a growing need for food, agricultural systems must become more productive and more sustainable. Especially intensive cropping systems consume significant amounts of non-renewable inputs such as fertilizers, fuel and growing media containing peat. Chemical pesticides, applied to protect crops from biotic stresses and yield losses, cause environmental contamination. Meanwhile, organic materials are treated as waste, but have the potential to be re-used as fertilizer, as soil improver or as part of growing media. In this context, the paradigmatic shift in waste management initiated by the European Waste Framework Directive (Council Directive 2008/98/EC) enhances the possibility of continuing the life cycle of materials in the perspective of a circular economy, i.e. organic rich waste can be applied for agricultural beneficial use allowing organic matter and nutrient cycling, as alternatives to their landfilling or incineration (Alvarenga et al. 2017; Estrada De Luis and Gómez Palacios 2013; Manfredi and Pant 2013). Nowadays, a wide range of resources and products with the potential to overcome soil constraints, improve nutrient use efficiency or support plant health, is available for application in agriculture (Abbott et al. 2018).
Also micro-organisms have the potential to enhance plant growth and protect them against biotic and abiotic stresses (Reid and Greene 2012; Parnell et al. 2016; Kim and Anderson 2018). The presence of beneficial organisms in soil can be stimulated by organic amendments as well as biostimulants, or by inoculating microbial strains (Calvo et al. 2014; Abbott et al. 2018). Efficacy of all these products, separate or in combination, is often unclear and very much dependent on the environmental condition, soil type as well as the crop type. More experimental data on efficacy of these products to support all the different claims are needed (Abbott et al. 2018).
Long term experience is available with compost applications in agriculture and horticulture to improve soil quality and fertility (Martinez et al. 2006). Compost can also substitute peat in potting soil (Vandecasteele et al. 2018; Avilés et al. 2011). Moreover, compost is often mentioned for its disease suppressive effects against several foliar and soil-borne diseases (Termorshuizen et al. 2006; Noble and Coventry 2005; Bonanomi et al. 2010), but results can be variable and the effects are case-specific (Martinez et al. 2006).
More recently, positive effects of biochar as a soil amendment are appointed on plant growth and soil quality (Bonanomi et al. 2015; Glaser et al. 2002; Jeffery et al. 2011). Biochar is a relatively recalcitrant product generated through pyrolysis, i.e. a reductive thermal process carried out at temperatures from 300 to 850 °C, whereas input material as well as temperature and residence time are crucial for the quality of the resulting carbonized product (Someus and Pugliese 2018). A meta-analysis of 16 studies on biochar application to soil indicated that improved water holding capacity, liming effect and improved crop nutrient availability are the main causes for increased crop productivity (Jeffery et al. 2011). Biochar is also indicated as sustainable alternative for the use of peat in potting soil reducing the peat derived CO2 emissions (Blok et al. 2017; Kern et al. 2017). Some studies indicate the potential of biochar to reduce foliar diseases and pests (Kumar et al. 2018; Meller Harel et al. 2012; De Tender et al. 2016) as well as soil-borne plant pathogens (Jaiswal et al. 2017; Huang et al. 2015; Jaiswal et al. 2014). However, compared to compost, very few experimental studies on biochar controlling plant diseases are available (Bonanomi et al. 2015).
To create more sustainable growing systems for greenhouse crops, both compost and biochar can be added to potting soil to recycle nutrients and to substitute part of the peat. In addition, the impact of these amendments on disease suppressiveness is relevant, but the variability in disease control among different crops and/or systems will complicate the application. Addition of microbial inoculants, alone or in combination with the organic amendments, might provide more robust disease suppressive effects (Pugliese et al. 2011; Bonanomi et al. 2018). Especially artificial growing media (i.e. rockwool, perlite) and potting soil can benefit from introducing micro-organisms not yet present, since such media start with a low microbial biodiversity and lack plant associated microbes. Some studies demonstrated an increased disease suppression by the addition of biocontrol micro-organisms to compost (Postma et al. 2003; Trillas et al. 2006; Pugliese et al. 2011). Other studies tested the potential of biochar to be used as carrier for beneficial micro-organisms (Hale et al. 2015; Postma et al. 2013).
In the current research, disease suppressive effects of several composts and two biochar types were evaluated in four independent greenhouse experiments performed with young tomato plants in potting soil. All potting soil mixtures were tested without (i.e. healthy controls) and with the pathogen Pythium aphanidermatum inoculated to the potting soil prior to sowing. Pseudomonas chlororaphis strain 4.4.1, which previously successfully controlled P. aphanidermatum (Postma et al. 2010), was inoculated to potting soil, biochar or tomato seeds, alone and in combination with the different organic amendments. Seed germination, number of healthy plants, plant growth and nutrient uptake were evaluated. Root colonization by the inoculated P. chlororaphis 4.4.1 strain was assessed by making use of a novel designed quantitative PCR system targeting the rpoD gene of P. chlororaphis 4.4.1.
Materials and methods
Growing medium and organic amendments
Potting soil was used as growing medium for all greenhouse experiments. This potting soil (product nr. 80,917, Lentse Potgrond BV, the Netherlands) consisted of 26% German/Irish fraction 0, 14% Baltic peat medium, 40% Swedish fine peat moss, 20% perlite, 0.5 kg m−3 PG-mix 12–14-24, 0.025 kg m−3 Superspoor MMS, and had a pH of 5.2.
Organic amendments (see overview in Table 1) were mixed through this potting soil. Two different types of biochar, animal-bone char (ABC) produced by Terra Humana ltd. (Biofarm Agri Research Station, Hungary) and plant-based biochar (PBC) (PYREG®) produced by Pyreg GmbH (Dörth, Germany), were applied in a dosage of 1% (weight/volume). ABC is made from food-grade animal bones at high carbonization temperature 850 °C and consists mainly of calcium phosphate. PBC is made from plant materials at a lower carbonization temperature, usually 450 °C, having a high carbon content.
A Dutch green waste compost (0–15 mm fraction) produced by Van Iersel Biezenmortel BV (the Netherlands) coded CONL was used in all experiments in a 10% dosage (volume/volume). Sterilized CONL compost was used for assessment of microbial effects on P. aphanidermatum disease suppression in the 2015 experiment (sterilization by gamma-radiation at 60 kGray; Synergy Health Ede, the Netherlands). Seven composts from other origins were included in the experiments in 2014 and 2015 (dosage 10% volume/volume).
Detailed chemical analysis of the organic amendments, including organic pollutants such as PAHs (polycyclic aromatic hydrocarbons) and heavy metals, were performed by Wessling (Hungary Kft., Budapest). A selection of the characteristics, focussing on the plant nutritional value, is summarized in Table 1.
Pathogen inoculum
The pathogen Pythium aphanidermatum 89 is causing pre- and post-emergence damping off in different plant species, including tomato seedlings. The strain was stored in liquid nitrogen. The pathogen was grown in 100 ml Erlenmeyer flasks with 25 ml liquid V8 medium, containing 5 ml V8 vegetable juice (Campbell Foods, Puurs, Belgium), 75 mg CaCO3, and 20 ml demineralized water (Postma et al. 2010). Flasks were incubated in the dark without shaking at 25 °C for 8–10 days. After incubation, the mycelium was washed three times with 50 ml sterilised tap-water to remove the medium, and blended in a Warring blender. Inoculum from one flask was used to infest 2.5 l growing medium, resulting in an inoculum density per experimental unit (pot) of approximately 105 colony-forming units (CFU) consisting of viable mycelium fragments and spores. Inoculum concentration was checked by plating serial dilutions on ¼ potato dextrose agar (PDA; 9.6 g PDA (Oxoid), 11.2 g agar per litre; 100 mg l−1 streptomycin and 10 mg l−1 tetracycline were added after autoclaving).
Bacterial antagonist P. chlororaphis 4.4.1
The bacterial strain inoculated to the different growing media in the experiments 2012, 2013, 2014 and 2015 was selected for its capacity to mobilize phosphorous (Postma et al. 2010) and its potential to control diseases in tomato plants (Postma et al. 2013). Strain 4.4.1 was originally isolated from an arable soil (Postma et al. 2008) and later identified as Pseudomonas chlororaphis subsp. aureofaciens based on sequencing of a 1500 bp fragment of the 16S rRNA gene (EMBL accession FR682804). The strain was maintained in freeze stock culture (−80 °C in 20% glycerol) and freshly cultured from frozen stocks prior to each experiment.
Inoculum was produced on R2A (Difco) plates at 25 °C. Bacterial cells were scraped from the plates and diluted in R2B (broth) to obtain the proper inoculum concentration. Ingredients of R2B per litre were 0.5 g proteose peptone, 0.5 g casamino acids, 0.5 g soluble starch (Difco), 0.5 g dextrose, 0.3 g sodium-pyruvate, 0.3 g K2HPO4 and 0.05 g MgSO4 (Sigma). The inoculum density was checked during preparation by measuring the optical density of the suspension. Final concentration was assessed by plating serial dilutions of the inoculum on R2A plates in duplicate and counting colony-forming units (CFU) after 2–4 days incubation at 25 °C.
P. chlororaphis was also applied to the tomato seeds in the 2015 experiment following a procedure which has been used for other non-spore forming gram-negative bacteria (Abuamsha et al. 2011). The method was adapted slightly for treatment of tomato seeds. Seeds were gently stirred during 30 min in 5 ml suspension P. chlororaphis with 1 × 107 CFU ml−1 in MgSO4 (0.1 M l−1). Thereafter the excess of water was removed on filter paper. Colonization of the seeds with P. chlororaphis was determined in duplicate by plate counting. Ten seeds per replicate were homogenized in 1 ml Ringers solution in BIOREBA extraction bags. A dilution series was plated on R2A medium as described above.
Effect of potting soil treatments on Pythium suppression
The ability of P. chlororaphis and two organic amendments to control P. aphanidermatum in tomato seedlings was first tested in two independent greenhouse experiments (2012 and 2013) as described by Postma et al. (2013). Four growing media were tested: potting soil alone (Control), and potting soil mixtures with 1% animal-bone char (ABC), 10% compost (CONL), or 10% compost +1% ABC (CONL+ABC). The assay was performed in plastic pots (11 × 11 × 12 cm) with 500 cm3 growing medium (equivalent to 91 g moist product). P. chlororaphis was added as liquid formulation directly to the growing media or to the animal-bone char (ABC) particles prior to their addition to the growing media. All inoculations of P. chlororaphis in the growing media were designed to have a final concentration of 106 cells g−1 potting soil. The microbial enriched ABC, used as a solid formulation, was directly applied to the potting soil without drying and contained 108 cells P. chlororaphis g−1 ABC. All growing media were also tested without P. chlororaphis and all treatments were assessed with and without pathogen inoculum. P. chlororaphis and pathogen inoculum were mixed gently through the potting soil mixtures, pots were filled with these mixtures and placed in the greenhouse at 25/20 °C d/n with 16 h light. Pots were covered with a plastic sheet to avoid evaporation. After 2 days incubation, 10 tomato seeds cultivar Pronto (De Ruiter Seeds, Bergschenhoek, The Netherlands) were sown per pot. Germination of seeds and post-emergence damping-off symptoms were monitored for a period of 21 days.
Two additional greenhouse experiments were performed in 2014 and 2015 to repeat the effect of ABC and the CONL compost on disease suppression and to evaluate an additional type of biochar, as well as seven other compost types. The bacterial antagonist P. chlororaphis was added directly to the growing media and in 2015 also as seed treatment. The assay was performed similar as described above, with few changes. All amendments were mixed with the potting soil mixtures 7 days before sowing and incubated in bags at 25/20 °C (day/night). Three (2014) or two (2015) days before sowing the pathogen and antagonist, or equal amounts of water in case of the control, were added and mixed through the soil. The concentration of P. chlororaphis when added as liquid to the growing media was adjusted to a 10 times higher final density then in the previous experiments, i.e. 107 cells g−1 soil. In 2015, the antagonist was also applied as a seed treatment (method is described above) at the day of sowing. On average 2.4 × 103 CFU seed−1 were present after seed treatment. In the presence of the pathogen 10 tomato seeds were sown per pot and germination of seeds and post-emergence damping-off symptoms were monitored for a period of 21 days. In the treatments without the pathogen, five seeds per pot were sown to determine plant biomass and nutrient uptake 25 days after sowing; seed germination and number of healthy plants were monitored as well.
All experiments were carried out with four replicates in a randomized block design with one pot of each treatment per block.
Nutrient analysis
Above ground parts of healthy plants were harvested after 25 days of growth and dried at 70 °C to determine fresh and dry weight (2014 and 2015). Thereafter the dried plant material was grinded, and after extraction total N, P and K concentrations were analysed at CBLB (ISO-17025 accredited lab, Wageningen, the Netherlands).
Rhizosphere sampling and DNA extraction
Presence and localization of P. chlororaphis 4.4.1 on the tomato roots was visualized at the end of the experiment in 2013. Roots of healthy plants were taken out of the soil, washed with tap water, excess of water was removed with filter paper and roots were pressed on square Petri dishes with agar medium (R2A). Roots were removed with forceps and the plates with imprints were incubated for 3 days at 25 °C. P. chlororaphis was recognized by its yellowish-orange colony pigmentation.
For quantitative detection of P. chlororaphis 4.4.1, tomato rhizosphere samples were harvested from treatments without Pythium inoculation in 2012, 2013 and 2015, respectively 21, 26 and 25 days after sowing. Roots with adhering potting soil were cut into pieces and stored at −80 °C. For each sample 0.4 g of root pieces with adhering potting soil was used for DNA extraction with a PowerMag® Soil DNA Extraction kit, optimized for KingFisher (Mo Bio Laboratories Inc., Carlsbad, USA). The manufacturer’s protocol was adapted to the combination of roots and potting soil. Lysis was done in 5 ml tubes to accommodate 2.5 times more lysis buffer per sample and all following steps were subsequently adapted resulting in 200 μl eluted DNA for each sample after KingFisher wash-steps. DNA of all samples was quantified with a PicoGreen assay (Quant-iT PicoGreen dsDNA Assay kit, Invitrogen) on Infinite M200pro (Tecan Group Ltd) according to the manufacturers’ instructions. All samples were diluted to 5 ng μl−1 DNA, prior to quantitative PCR.
Primer design of P. chlororaphis 4.4.1 and quantification of microbial populations in rhizosphere soil
A strain specific primer system was designed for P. chlororaphis strain 4.4.1. For that purpose, type strains of P. chlororaphis subsp. aureofaciens, subsp. aurantiaca, subsp. chlororaphis and P. corrugata were used, as well as a selection of strains available from our collection that were expected to be P. chlororaphis based on partial 16S sequences and their production of green to orange exo-pigments during growth (Table 2). Also three other Pseudomonas spp. strains and two less related bacterial strains were included. P. chlororaphis strain 4.4.1, type strain P. chlororaphis subsp. aureofaciens DSM 6698, and strains 10.2.5 and 11.4.2 appeared to be identical in REP-PCR (repetitive extragenic palindromic-PCR) (Rademaker et al. 1998) fingerprint analysis. Consequently, no suitable specific band allowing the design of primers and probes based on a strain specific sequence as described by Nijhuis et al. (2010) was available.
The strategy for designing specific primers for P. chlororaphis 4.4.1 detection was to target the rpoD (RNA polymerase sigma factor) gene, which is most discriminating out of four protein-coding genes studied in the genus Pseudomonas (Mulet et al. 2010). The rpoD fragments of P. chlororaphis 4.4.1 and the three closely related strains were amplified with a nested approach (Yamamoto et al. 2000). For the first PCR the 70F/70R primer set was used, followed by a second PCR on the 70F/70R amplicons with 70Fs/70Rs (Yamamoto et al. 2000) and PsEG30F/PsEG790R (Mulet et al. 2009). Sequences were aligned using CLC Main Workbench (www.clcbio.com) with 146 rpoD sequences for the Pseudomonas genus retrieved from NCBI database to visualize possible interesting areas in the sequence for primer and probe design targeting subspecies level. The NCBI Primer-Blast tool (Ye et al. 2012) was used to find suitable primers in these areas. The thus designed rpoD primer set, as well as another rpoD primer set developed for P. chlororaphis strain 30–84 (Wang et al. 2012), were evaluated for their specificity with all strains listed in Table 2, i.e. band intensity, size, and presence of by-products were taken into account. Then, the performance of the selected rpoD primer set was tested in qPCR (SybrGreen assay, described below) on dilution series of P. chlororaphis 4.4.1 (2–0.0002 ng μl−1 DNA) compared to the strains listed in Table 2 (0.2 ng μl−1 DNA) with two primer concentrations (300 and 600 nM).
P. chlororaphis 4.4.1 populations were quantified in the rhizosphere samples with qPCR using the newly designed rpoD primer set. In addition, more general primer sets to quantify Pseudomonas spp., total bacteria and fungi were included for the same samples. Used primer sets and references are listed in Table 3. qPCR was performed as a SybrGreen assay with Sybr® Premix Ex Taq (Takara Bio Inc., Otsu, Japan) to quantify P. chlororaphis 4.4.1 and total bacteria. For the SybrGreen assays each 25 μl reaction contained 2 μl (= 10 ng DNA) of sample, 12.5 μl Sybr Premix Ex Taq (2x), 0.5 μl of ROX reference Dye and either 300 nM for the rpoD primers or 600 nM for the total Bacteria primers. Premix Ex Taq (Takara Bio Inc., Otsu, Japan) was used for the Pseudomonas and fungi TaqMan assays. For each TaqMan PCR reaction of 25 μl, 2 μl sample was mixed with 23 μl of reaction mix containing 12.5 μl of Premix Ex Taq 2x, 0.5 μl of ROX reference Dye, 100 nM FAM-labelled hydrolysis probe, and 600 nM for the Pseudomonas primers or 300 nM for fungal primers.
In every 96-well plate, negative controls (water) and a standard curve were included at least twice. In the qPCRs for P. chlororaphis 4.4.1, Pseudomonas spp. and total bacteria, the standard curves consisted of a freshly prepared 10-fold serial dilution series of P. chlororaphis 4.4.1 DNA, prepared from a suspension with optical density of 0.1 (≈8.0 log CFU ml−1). CFU number in the original cell suspension was assessed by enumeration on R2A plates in duplicate. This calibration series was used to transform resulting Cq (quantification cycle) values to log CFU g−1 rhizosphere. For the total fungi TaqMan assay, a freshly prepared 10-fold serial dilution series with known amount of fungal DNA was included, ranging from 5 to 0.00005 ng μl−1 of Fusarium nivale DNA. This calibration series was used to transform resulting Cq values to log fg DNA g−1 rhizosphere. Since fungi consist of different propagules (spores and mycelia) DNA concentrations cannot be transformed into CFU values. Standard curves for all qPCRs were calculated with linear regression by plotting the Cq value against the log CFU g−1 rhizosphere or log fg DNA g−1 rhizosphere for fungi.
All qPCRs were carried out in an Applied Biosystems 7500 Real-Time PCR system (Applied Biosystems) using the standard program, except for PCRs with primer set for total bacteria where a lower annealing temperature of 53 °C was used. For all SybrGreen assays dissociation curves were included.
Statistical analysis
Analyses of variance (ANOVA) and regression analysis were carried out with the statistical program GenStat release 18.1 (Rothamsted Experimental Station, Harpenden, UK). All plant experiments were carried out in a randomized block design with four replications (independent pots) per treatment. Microbial numbers were analysed after logarithmic (10log(x)) transformation. Multi-factor analyses were performed with type of amendment, P. chlororaphis inoculation and year as factors. After ANOVA, least significant differences (LSD) were calculated at a significance level of P = 0.05. The percentage of healthy plants in the Pythium infected treatments was correlated with the qPCR detected P. chlororaphis populations in the rhizosphere (log scale) by regression analysis for the different organic amendments.
Results
Seed germination
In the absence of P. aphanidermatum, on average 96, 98, 97 and 98% of the tomato seeds germinated and remained healthy in the four independent experiments in respectively 2012, 2013, 2014 and 2015. The bacterial antagonist P. chlororaphis 4.4.1 did not significantly improve or reduce germination in the absence of the pathogen P. aphanidermatum. Also the applied dosages of the organic amendments did not significantly influence germination of the seeds; except for two compost batches in the 2014 experiment COSPBS and COSPBD that seriously reduced seed germination, i.e. germination rates were 68 and 73% respectively. These composts were prepared from green waste with manure for fertilizer purpose and appeared to be phytotoxic when added in a 10% dosage to potting soil. Therefore, these composts were not used in further experiments to evaluate the effect of amendments on Pythium control.
Pythium control by organic amendments
Inoculation with the pathogen P. aphanidermatum resulted in significant losses of tomato plants during and after seed germination, due to respectively pre- and post-emergence damping off. Only 43 to 68% of the tomato plants remained healthy in the control treatments with unamended potting soil when the pathogen was added in the experiments of the different years, whereas the pathogen-free treatments had 96 to 98% healthy plants.
The experiments in 2012 and 2013 did not show any significant interaction between treatments and years, allowing the presentation of average values of both experiments (Fig. 1a). Potting soil amended with compost (CONL) resulted in a significantly greater number of healthy plants (85%) compared to the non-amended control potting soil (50%). Amendment with ABC (without microbial inoculant) did not significantly increase the number of healthy plants as compared to the control, or when added to the treatment with compost. Results were based on one single dosage applied to the potting soil, i.e. 10% compost and 1% ABC.
Healthy tomato plants in potting soil (Control) and potting soil amended with animal-bone char (ABC), plant-based biochar (PBC), compost (CONL) or a combination of compost with ABC (CONL+ABC), 21 days after inoculation with Pythium aphanidermatum. All treatments were without (No) or with (Pc) Pseudomonas chlororaphis 4.4.1 inoculation. Upper fig. (a): average of experiments in 2012 and 2013 with a least significant difference (LSD) of 16.5 at P = 0.05. Lower fig. (b): average of experiments in 2014 and 2015 with LSD of 18.6 at P = 0.05. Bars with different letters are significantly different. (N = 8)
In the experiments 2014 and 2015, amendments with compost (CONL), as well as animal-bone char (ABC) and plant-based biochar (PBC) were tested in the same dosage as before. Since there was no significant interaction between these treatments and years, the average values of the two experiments are presented (Fig. 1b). Again, CONL significantly increased the number of healthy plants, whereas ABC had no effect. The effect of PBC was intermediate and not significantly different from the control.
Also several other types of compost were evaluated in 2014 and 2015, which all demonstrated higher numbers of healthy plants compared to the unamended potting soil, and equally or more healthy plants than the CONL compost that was used in all experiments (Fig. 2; for results of 2014 see SUPPLEMENTARY Fig. 1). Surprisingly, also the sterilized CONL compost (CONL-st) enhanced the number of healthy plants compared to the control (Fig. 2).
Healthy tomato plants in potting soil (Control) and potting soil amended with animal-bone char (ABC), plant-based biochar (PBC), four different compost types (CO), and sterilized compost (CONL-st) in 2015, 21 days after inoculation with Pythium aphanidermatum. All treatments were without the antagonist (No) or with the bacterial antagonist Pseudomonas chlororaphis 4.4.1 inoculated to soil (Pc) or seed (Pc-seed). Least significant difference (LSD) is 17.8 at P = 0.05. Bars present mean values ± standard deviation. (N = 4)
Pythium control by bacterial antagonist P. chlororaphis 4.4.1
The antagonist P. chlororaphis was added in all experiments to each of the growing media. Different inoculation strategies were tested. Comparison of P. chlororaphis added as liquid formulation to potting soil or as solid formulation within the ABC particles in the experiments of 2012 and 2013 demonstrated that there was no difference in the effect due to the formulation (Fig. 1a); in both treatments (Control and ABC) P. chlororaphis inoculation resulted in approximately 80% healthy plants, compared to 50 and 56% healthy plants in the treatments without P. chlororaphis. In the treatments with compost, where the growing medium was already reducing the Pythium symptoms, P. chlororaphis did not further elevate the number of healthy plants; i.e. there was no additive effect of the antagonist combined with compost amendment (Fig. 1a).
Inoculation of P. chlororaphis as liquid formulation in experiments 2014 and 2015 had similar results (Fig. 1b) as the 2012 and 2013 experiments (Fig. 1a). Inoculation of P. chlororaphis significantly increased the number of healthy plants in de control potting soil as well as in the potting soil amended with ABC. Number of healthy plants were not further increased by inoculating P. chlororaphis in the treatments with PBC or compost that were already (partly) controlling the Pythium symptoms (Fig. 1b). Inoculation of P. chlororaphis to all other compost amendments had similar effects as the compost amendment itself (Fig. 2 and SUPPLEMENTARY Fig. 1); i.e. there was no additive effect of the antagonist combined with any type of compost amendment.
In 2015, P. chlororaphis was also introduced through seed treatment (Pc-seed). Although the antagonist inoculum dosage was very low (2.35 × 103 CFU seed−1), significantly more healthy plants were present after seed treatment with P. chlororaphis in the ABC and PBC amended potting soil compared to the same growing media without P. chlororaphis inoculation (Fig. 2). The numbers of healthy plants in the control potting soil were higher at a significance level of P = 0.10. In general, the seed treatment with P. chlororaphis was as effective in controlling Pythium as the addition of the antagonist to the growing media (Fig. 2).
Plant growth and nutrient uptake
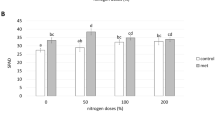
The effect of the different organic amendments and P. chlororaphis on plant biomass and nutrient uptake by the tomato plants was measured in the experiments 2014 and 2015 using healthy plants in the absence of the pathogen. There was no significant effect due to inoculation with P. chlororaphis compared to the non-inoculated treatments, therefore average values of treatments with and without P. chlororaphis are presented in Fig. 3. Dry weight of the plants (Fig. 3a) and total nitrogen (N-total) (Fig. 3b) were similar for most of the treatments. Only the compost treatment in 2014 had a higher level of N-total than the control potting soil (Fig. 3b). However, there were important differences in P and K content of the plants. P levels in the plants were twice as high in the treatments with ABC and compost compared to the control (Fig. 3c). PBC was intermediate, but results differed in both years. K levels in the plants were increasing in the order: control, ABC, PBC and compost (Fig. 3d).
Plant biomass (dry weight) and nitrogen (N-total), phosphorous (P) and potassium (K) uptake by 25-day old tomato plants in 2014 and 2015. Average data for treatments with and without Pseudomonas chlororaphis 4.4.1 inoculation. Least significant differences (LSD) at P = 0.05 are 0.69, 0.22, and 1.59 for respectively N-total, P, and K uptake, whereas dry weight did not significantly differ between the treatments. Bars with different letters are significantly different. (N = 8)
Detection method for P. chlororaphis 4.4.1
The rpoD sequences of P. chlororaphis 4.4.1 were found to be identical to the sequences of 10.2.5, 11.4.2 and the type strain DSM 6698 (NCBI accession numbers of rpoD sequences of the four strains are MK574038, MK574039, MK574040 and MK574041, respectively). The fragment size of 728 bp, representing these four strains, was used for primer design with the NCBI Primer-Blast tool (Ye et al. 2012). The designed primers matched 100% to P. chlororaphis subsp. aureofaciens strains LMG1245T and NCIMB 9030 entries in NCBI database (FN554453 and AB039554, respectively) which are other collection numbers for the type strain P. chlororaphis subsp. aureofaciens DSM 6698. In the forward primer sequence, one mismatch was found with P. chlororaphis subsp. aureofaciens 30–84 and LMG5832 and in both primers for P. chlororaphis subsp. aureofaciens NCIMB 9265 (NCBI entries HE586461 and JN397563, AB039555, respectively). PCR tests with this rpoD primer set resulted in strong single bands of the expected size with the target strain 4.4.1, as well as the strains with identical rpoD sequences DSM 6698, 10.2.5, 11.4.2, and the two other P. chlororaphis type strains DSM 19603, DSM 50083 and strain 1110–027, but not with the other Pseudomonas species or the Burkholderia and Lysobacter strains. The simultaneously tested rpoD primer set for P. chlororaphis strain 30–84 (Wang et al. 2012) reacted with the same strains as our rpoD primer set, but also with P. corrugata type strain DSM 7228 and P. fluorescens strain 5 (Table 2).
In the qPCR tests, Cq values and estimated melting temperatures (Tm) in dissociation curves were similar for our target strain 4.4.1, DSM 6698, 10.2.5 and 11.4.2. Similar Cq values and Tm slightly lower but within −0.5 °C range were found for DSM 19603 and DSM 50083. For strain 1110–027, Cq values shifted up with 2 cycles and Tm was 1 °C lower. All other strains showed negative reactions: Cq values increased 8 to 15 cycles and Tm dropped with −1.5 to −13 °C. As a comparison, in negative water controls Cq values increased 16 to 17 cycles and Tm dropped with −13 °C.
For all performed qPCR assays, the slopes of standard curves were between of −3.1 and − 3.6, E values were above 99% and all R2 values were above 0.99, which is in the same range as recommended by Smith and Osborn (2009) and Bustin et al. (2009). For our rpoD primer set, qPCR standard curves with 300 nM matched the above mentioned criteria, whereas standard curves with 600 nM did not. Therefore, primer concentration of 300 nM was used in the rpoD qPCRs.
Although the developed qPCR system was quite specific, a strictly strain specific detection of P. chlororaphis 4.4.1 based on rpoD sequences was not feasible. The rpoD based qPCR targeted a subgroup of P. chlororaphis, and results on presence of the inoculated strain should therefore be compared with non-inoculated treatments to take background levels of naturally present P. chlororaphis strains into account.
Rhizosphere colonization by P. chlororaphis and other microbial populations
Healthy plants without Pythium inoculation of the greenhouse experiments in 2012, 2013 and 2015 were used to evaluate the rhizosphere colonization in different amended growing media, with and without P. chlororaphis inoculation. Qualitative tests in 2013, by making an imprint of washed root system on nutrient medium, demonstrated that bacterial colonies (recognized by its yellowish-orange colony pigmentation) were all along the roots (data not shown). Quantitative detection of P. chlororaphis populations with qPCR demonstrated that P. chlororaphis could establish in the rhizosphere up to 3 weeks after inoculation: all inoculated treatments had higher P. chlororaphis values compared to the non-inoculated (No) treatments (Pc-subgroup in Table 4). Average values of P. chlororaphis after inoculation were above 104 cells g−1 rhizosphere, and were 4 to 100 times higher than without inoculation of the antagonist in the different years. The final concentration of P. chlororaphis on the tomato roots was similar in the different experiments, although the inoculum density was 10 times higher in 2015 as compared to 2012 and 2013. The different organic amendments appeared to have no clear effect on the root colonization by P. chlororaphis (see SUPPLEMENTARY Table 1 and 2 for the results of the individual treatments).
Inoculation of P. chlororaphis through seed treatment resulted in an average of 4 × 103 cells g−1 rhizosphere, which was 7 times higher than without inoculation (Table 4). These lower population levels are probably due to the rather low inoculum dosage used for seed inoculation resulting in 2.4 × 103 cells seed−1 at the start of the experiment.
Increase of the Pseudomonas spp. counts after inoculation with the biocontrol strain can be attributed to the inoculation with P. chlororaphis 4.4.1. In general the Pseudomonas numbers surpassed the numbers of the Pc-subgroup, and in the treatments where P. chlororaphis 4.4.1 was inoculated, the share of Pc-subgroup in the Pseudomonas spp. numbers was higher (Table 4). Pseudomonas populations in the rhizosphere of the non-inoculated treatments were in general lowest in the control potting soil without organic amendment, but this was not a consistent and significant difference (SUPPLEMENTARY Table 1 and 2 show the results of the individual treatments).
Total bacterial and total fungal numbers were not influenced by the inoculation with P. chlororaphis (Table 4). Moreover, bacterial and fungal populations were not systematically influenced by the organic amendments. Only in part of the compost treatments elevated numbers of bacteria or fungi were present, but this was not consistent (SUPPLEMENTARY Table 1 and 2).
Correlation between plant health and P. chlororaphis populations
Taking all P. chlororaphis population measurements and plant health results of the 2012, 2013 and 2015 experiments into account, there was no significant correlation (correlation coefficient of 0.23). Therefore, regression analysis was performed per type of growing medium. In the unamended potting soil, addition of P. chlororaphis resulted in higher populations of the inoculated antagonist (qPCR detection of Pc-subgroup) as well as in more healthy plants; a significant linear correlation could be fitted with regression analysis. The % of healthy plants corresponded with 29.2 + 10.2 x log value of the Pc-subgroup (variance accounted for was 22.7%; standard error of observations was estimated to be 17.9). In the compost amended potting soil, addition of P. chlororaphis also resulted in higher populations of the inoculated antagonist (qPCR detection of Pc-subgroup), but there was no increase of healthy plants. Consequently, there was no significant linear correlation. Potting soil amended with ABC followed the same trend as the unamended potting soil. From the other growing media the number of data were too few for proper regression analysis. For visualization of the correlation between plant health and P. chlororaphis populations see SUPPLEMENTARY Fig. 2.
Discussion
Several types of compost and two biochars were mixed with potting soil in four independent greenhouse experiments. Their efficacy on plant growth and plant health was tested alone or in combination with a biocontrol strain. Several of the organic amendments as well as the biocontrol strain P. chlororaphis 4.4.1 repeatedly protected young tomato plants against infection with the plant pathogen P. aphanidermatum. The focus of this research was on disease suppression, but in addition the potential of the organic products for nutrient cycling was assessed by measuring growth and nutrient uptake by the young tomato plants.
Compost amended in a 10% dosage to the potting soil, significantly reduced the number of infected tomato plants in the presence of the pathogen P. aphanidermatum compared to unamended potting soil in all four experiments. Surprisingly, all green-waste composts tested reduced Pythium infection. Generally, the effect of compost on suppression of plant pathogens is indicated to be dependent on their physical, chemical and biological characteristics (Termorshuizen et al. 2006; Avilés et al. 2011). Several mechanisms of disease suppression by compost are described. Microbial enrichment due to the compost is generally mentioned to be involved (Avilés et al. 2011). In our study, this mechanism is unlikely, since sterilized compost was also effective in reducing the infection. Moreover, the total number of bacteria and fungi as well as the pseudomonas populations in the rhizosphere of suppressive and non-suppressive treatments did not clearly differ (qPCR detection). The fact that eight different types of green-waste compost were all reducing disease incidence indicates that specific micro-organisms do not play a role. Pythium spp. are described as highly sensitive to nutrient competition and antibiosis, and most likely the green-waste composts have induced an elevated microbial activity contributing to general disease suppression (Avilés et al. 2011). Also some other studies demonstrated such a general suppressive effect of compost against Pythium spp. when amended to peat-like growing media (Pane et al. 2011; Alfano et al. 2011; Vestberg et al. 2014).
Application of biochar in agricultural soils to improve soil quality and to enhance plant growth is a relatively new development, and an increasing number of publications is available only from the last 15 to 20 years (Bonanomi et al. 2015; Glaser et al. 2002; Jeffery et al. 2011). In our experiments we applied two very distinct types of biochar in a 1% dosage to potting soil, i.e. a plant-based biochar and a biochar produced from animal by-products. These products are very different in composition and are generally applied for different purposes. Plant-based biochar (PBC) is carbon-rich and aimed for soil improvement, whereas animal bone char (ABC) is rich in calcium phosphate and useful as P fertilizer. In the current research we focussed on disease suppressive potential of these products alone or in combination with a biocontrol strain. The effect of PBC on disease suppression was intermediate to compost and the control potting soil. ABC did not show any increase of Pythium disease suppression in all four experiments. Previous experiments showed little effect of ABC on disease suppression of the soil-borne pathogens Pythium and Fusarium when the material was used without microbial additions (Postma et al. 2013). Some studies demonstrated the potential of plant-derived biochar types to reduce foliar diseases and pests (Kumar et al. 2018; Meller Harel et al. 2012; De Tender et al. 2016), as well as few soil-borne plant pathogens (Jaiswal et al. 2017; Huang et al. 2015; Jaiswal et al. 2014). Often the mechanism claimed is induced resistance (Bonanomi et al. 2015). However, disease suppressiveness is in general crop- and pathogen-specific and studies with positive effects of biochar on suppressiveness of Pythium spp. have not been found (Dorais et al. 2016; Gravel et al. 2013). Also the applied concentration of biochar is relevant. Higher dosages of biochar might be more effective, but some studies demonstrate negative effects on disease suppression when e.g. 3% or more biochar was applied (Frenkel et al. 2017).
A complementary strategy to enhance disease suppression is to add biocontrol organisms. Here we applied the strain P. chlororaphis 4.4.1 that controlled Pythium as well as Fusarium in a previous study when it was applied with ABC as a carrier (Postma et al. 2013). The biocontrol strain was inoculated with different procedures into the different potting soil mixtures. The strain consequently protected young tomato plants against P. aphanidermatum infection in the control potting soil. It also significantly enhanced suppressiveness of the potting soil that was amended with 1% ABC. However, when the compost amendment increased suppressiveness of the potting soil mixture, the biocontrol strain did not result in elevated numbers of healthy plants; i.e. there was no additive effect when combining the biocontrol strain P. chlororaphis with compost.
Efficacy of the biocontrol strain was independent from the inoculation procedure. Adding P. chlororaphis as liquid formulation to the growing medium, as well as seed inoculation were effective. Interestingly, P. chlororaphis could also be applied as a solid formulation with ABC as a carrier.
Seed inoculation is a good strategy to overcome the limitations of short shelf-life of non-spore forming bacteria. The applied seed inoculation procedure resulted in rather low numbers of P. chlororaphis, i.e. 2.4 × 103 cells seed−1. Nevertheless, protection of tomato plants against Pythium symptoms was significant. Studies on inoculating oil seed rape seeds with P. chlororaphis MA 342 demonstrated that time and inoculum concentration both influenced the bacterial concentration on the seeds (Abuamsha et al. 2011). Since treatment time of tomato seeds had to be shorter for proper seed germination than for oil seed rape (30 min instead of 4 h), higher cell densities in the inoculum suspension are advised to further optimize the inoculation procedure for tomato seeds.
Survival and root colonization of biocontrol organisms is a key factor for efficacy (Lugtenberg and Kamilova 2009). The imprint of the washed root system on nutrient medium demonstrated that colonies of the inoculated strain were along the entire root system. This confirms the root colonizing capacity of P. chlororaphis 4.4.1 which was demonstrated previously by plate counts using an antibiotic resistant mutant (Postma et al. 2013). However, a molecular detection method is more appropriate for quantitative detection of the wild type strain in the rhizosphere of different growing media. Therefore, a primer set was developed based on the rpoD protein-coding gene, which shows proper discrimination in the Pseudomonas genus (Mulet et al. 2010) and is present as single copy allowing quantitative detection (Mulet et al. 2009). P. chlororaphis was detected at low levels as a background in the treatments that were not inoculated with our strain, since the designed primer set targets a subgroup of P. chlororaphis. However, when P. chlororaphis 4.4.1 was inoculated numbers were significantly higher, indicating the usefulness of the rpoD primer set for the quantitative detection of P. chlororaphis 4.4.1. The root colonizing capacity of P. chlororaphis 4.4.1 was confirmed once more with this novel qPCR method, i.e. P. chlororaphis populations in the rhizosphere were 4 to 100 fold higher 3 weeks after the biocontrol strain was inoculated to soil or seeds as compared to non-inoculated treatments. The strain colonized the roots even when it was inoculated locally (on seeds) at a low dosage (2.4 × 103 cells seed−1).
Compost and biochar are generally applied for their fertilizer effect (Abbott et al. 2018). In our study we used fertilized potting soil to avoid interactions with disease infection while testing the disease suppressive effects, thus nutrients were not a limiting factor during tomato growth up to 3 weeks. As a result, plant growth (expressed as dry weight) of the healthy plants without Pythium inoculation was similar for all treatments. Nevertheless, nutrient uptake by the tomato plants was influenced by the organic amendments. Potassium (K) levels in the plant increased most distinct by the compost amendment, followed by PBC and ABC amendments. Phosphorous (P) levels increased by compost and ABC, and to a lesser extent by PBC. Here we should remark that the compost dosage was 10 times higher than the applied dosage of ABC and PBC. Since the global reserves of economically recoverable P will be depleted when increasing amounts of mineral P are used (Cordell et al. 2009), combined with the fact that mineral P is containing cadmium and uranium (Schnug and Lottermoser 2013), recycling P-rich organic materials is of great importance. ABC can be considered as a safe P fertilizer due to its purity and practically absence of contaminants (Someus and Pugliese 2018). The uptake of P was not improved by inoculating P. chlororaphis, although this strain was selected for its P-solubilising capacity (Postma et al. 2010). P availability of ABC depends on soil acidity (Warren et al. 2009) and the product formulation (Someus and Pugliese 2018). Since potting soil has a pH 5.2 and tomato roots exudate organic acids (Kamilova et al. 2006), the inoculated P. chlororaphis probably had no additional effect on P solubilisation under these conditions. To evaluate the fertilizer effect of the different organic amendments, nutrient poor soils or growing media in combination with longer growing periods of the crops must be tested, preferably under realistic growing conditions.
Our results demonstrate the potential of the assessed organic amendments to enhance sustainability of growing media by (i) increasing disease suppressiveness of the growing medium, (ii) re-use of nutrients present in the applied organic products, as well as (iii) to replace part of the peat by other organic materials. In addition, inoculating seed or growing medium with the biocontrol strain P. chlororaphis enhanced Pythium control in the susceptible growing media.
The evaluated products have very different characteristics, and are targeted for different purposes. The challenge in designing more sustainable systems is the proper application and combination of these products. Although inoculating the biocontrol strain P. chlororaphis into growing medium that became suppressive by compost amendment did not further enhance Pythium suppressiveness, such a combination can improve the robustness of the system, when the biocontrol strain is effective against other diseases as well. ABC with its high P content is a suitable organic fertilizer, meanwhile due to its porous structure it is an interesting carrier for micro-organisms. Enrichment with the biocontrol bacterium demonstrated that the product will become suppressive to plant diseases. PBC being a carbon rich product, is a soil improver with water and nutrient retention capacity. Due to its slow degradability, it can also be used in growing media to replace peat. In all cases, the recycled organic products should be targeted for agricultural purposes and meet all safety criteria.
References
Abbott, L. K., Macdonald, L. M., Wong, M. T. F., Webb, M. J., Jenkins, S. N., & Farrell, M. (2018). Potential roles of biological amendments for profitable grain production – A review. Agriculture, Ecosystems and Environment, 256, 34–50. https://doi.org/10.1016/j.agee.2017.12.021.
Abuamsha, R., Salman, M., & Ehlers, R.-U. (2011). Improvement of seed bio-priming of oilseed rape (Brassica napus ssp. oleifera) with Serratia plymuthica and Pseudomonas chlororaphis. Biocontrol Science and Technology, 21(2), 199–213.
Alfano, G., Lustrato, G., Lima, G., Vitullo, D., & Ranalli, G. (2011). Characterization of composted olive mill wastes to predict potential plant disease suppressiveness. Biological Control, 58(3), 199–207. https://doi.org/10.1016/j.biocontrol.2011.05.001.
Alvarenga, P., Palma, P., Mourinha, C., Farto, M., Dôres, J., Patanita, M., Cunha-Queda, C., Natal-da-Luz, T., Renaud, M., & Sousa, J. P. (2017). Recycling organic wastes to agricultural land as a way to improve its quality: A field study to evaluate benefits and risks. Waste Management, 61, 582–592. https://doi.org/10.1016/j.wasman.2017.01.004.
Avilés, M., Borrero, C., & Trillas, M. I. (2011). Review on compost as an inducer of disease suppression in plants grown in soilless culture. Dynamic Soil Dynamic Plant, 5(Special Issue 2), 1–11.
Bergmark, L., Poulsen, P. H. B., Al-Soud, W. A., Norman, A., Hansen, L. H., & Sørensen, S. J. (2012). Assessment of the specificity of Burkholderia and pseudomonas qPCR assays for detection of these genera in soil using 454 pyrosequencing. FEMS Microbiology Letters, 333(1), 77–84. https://doi.org/10.1111/j.1574-6968.2012.02601.x.
Blok, C., van der Salm, C., Hofland-Zijlstra, J., Streminska, M., Eveleens, B., Regelink, I., et al. (2017). Biochar for horticultural rooting media improvement: Evaluation of biochar from gasification and slow pyrolysis. Agronomy, 7(6), 1–23.
Bonanomi, G., Antignani, V., Capodilupo, M., & Scala, F. (2010). Identifying the characteristics of organic soil amendments that suppress soilborne plant diseases. Soil Biology and Biochemistry, 42(2), 136–144.
Bonanomi, G., Ippolito, F., & Scala, F. (2015). A “black” future for plant pathology? Biochar as a new soil amendment for controlling plant diseases. Journal of Plant Pathology, 97(2), 223–234.
Bonanomi, G., Lorito, M., Vinale, F., & Woo, S. L. (2018). Organic amendments, beneficial microbes, and soil microbiota: Toward a unified framework for disease suppression. Annual Review of Phytopathology, 56(1), 1–20. https://doi.org/10.1146/annurev-phyto-080615-100046.
Bustin, S. A., Benes, V., Garson, J. A., Hellemans, J., Huggett, J., Kubista, M., Mueller, R., Nolan, T., Pfaffl, M. W., Shipley, G. L., Vandesompele, J., & Wittwer, C. T. (2009). The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clinical Chemistry, 55(4), 611–622. https://doi.org/10.1373/clinchem.2008.112797.
Cai, G. H., Bröms, K., Mälarstig, B., Zhao, Z. H., Kim, J. L., Svärdsudd, K., et al. (2009). Quantitative PCR analysis of fungal DNA in Swedish day care centers and comparison with building characteristics and allergen levels. Indoor Air, 19(5), 392–400. https://doi.org/10.1111/j.1600-0668.2009.00600.x.
Calvo, P., Nelson, L., & Kloepper, J. W. (2014). Agricultural uses of plant biostimulants. Plant and Soil, 383(1–2), 3–41. https://doi.org/10.1007/s11104-014-2131-8.
Cordell, D., Drangert, J. O., & White, S. (2009). The story of phosphorus: Global food security and food for thought. Global Environmental Change, 19(2), 292–305.
Council Directive (2008)/98/EC. of 19 November 2008 on waste and repealing certain Directives. OJ L 312, 22.11.2008, p. 3–30.
De Tender, C. A., Debode, J., Vandecasteele, B., D'Hose, T., Cremelie, P., Haegeman, A., et al. (2016). Biological, physicochemical and plant health responses in lettuce and strawberry in soil or peat amended with biochar. Applied Soil Ecology, 107, 1–12. https://doi.org/10.1016/j.apsoil.2016.05.001.
Dorais, M., Martinez, C., Diop, M., Thériault, M., Ménard, C., & Pepin, S. (2016). Assessing the potential of biochar as a growing media component for potted plants. Acta Horticulturae, (1137), 19–26. https://doi.org/10.17660/ActaHortic.2016.1137.3.
Estrada De Luis, I. B., & Gómez Palacios, J. M. (2013). European Union (EU) end of waste regulation: Requirements for input materials and compost quality for sludge and other biodegradable wastes. Journal of Residuals Science and Technology, 10(3), 139–146.
Fierer, N., Jackson, J. A., Vilgalys, R., & Jackson, R. B. (2005). Assessment of soil microbial community structure by use of taxon-specific quantitative PCR assays. Applied and Environmental Microbiology, 71(7), 4117–4120. https://doi.org/10.1128/aem.71.7.4117-4120.2005.
Frenkel, O., Jaiswal, A. K., Elad, Y., Lew, B., Kammann, C., & Graber, E. R. (2017). The effect of biochar on plant diseases: What should we learn while designing biochar substrates? Journal of Environmental Engineering and Landscape Management, 25(2), 105–113. https://doi.org/10.3846/16486897.2017.1307202.
Glaser, B., Lehmann, J., & Zech, W. (2002). Ameliorating physical and chemical properties of highly weathered soils in the tropics with charcoal - a review. Biology and Fertility of Soils, 35(4), 219–230. https://doi.org/10.1007/s00374-002-0466-4.
Gravel, V., Dorais, M., & Ménard, C. (2013). Organic potted plants amended with biochar: Its effect on growth and Pythium colonization. Canadian Journal of Plant Science, 93(6), 1217–1227. https://doi.org/10.4141/cjps2013-315.
Hale, L., Luth, M., & Crowley, D. (2015). Biochar characteristics relate to its utility as an alternative soil inoculum carrier to peat and vermiculite. Soil Biology and Biochemistry, 81, 228–235. https://doi.org/10.1016/j.soilbio.2014.11.023.
Huang, W. K., Ji, H. L., Gheysen, G., Debode, J., & Kyndt, T. (2015). Biochar-amended potting medium reduces the susceptibility of rice to root-knot nematode infections. BMC Plant Biology, 15(267), 1–15. https://doi.org/10.1186/s12870-015-0654-7.
Jaiswal, A. K., Elad, Y., Graber, E. R., & Frenkel, O. (2014). Rhizoctonia solani suppression and plant growth promotion in cucumber as affected by biochar pyrolysis temperature, feedstock and concentration. Soil Biology and Biochemistry, 69, 110–118. https://doi.org/10.1016/j.soilbio.2013.10.051.
Jaiswal, A. K., Elad, Y., Paudel, I., Graber, E. R., Cytryn, E., & Frenkel, O. (2017). Linking the belowground microbial composition, diversity and activity to soilborne disease suppression and growth promotion of tomato amended with biochar. Scientific Reports, 7(44382), 1–17. https://doi.org/10.1038/srep44382.
Jeffery, S., Verheijen, F. G. A., van der Velde, M., & Bastos, A. C. (2011). A quantitative review of the effects of biochar application to soils on crop productivity using meta-analysis. Agriculture, Ecosystems and Environment, 144(1), 175–187. https://doi.org/10.1016/j.agee.2011.08.015.
Kamilova, F., Kravchenko, L. V., Shaposhnikov, A. I., Azarova, T., Makarova, N., & Lugtenberg, B. (2006). Organic acids, sugars, and L-tryptophane in exudates of vegetables growing on stonewool and their effects on activities of rhizosphere bacteria. Molecular Plant-Microbe Interactions, 19(3), 250–256.
Kern, J., Tammeorg, P., Shanskiy, M., Sakrabani, R., Knicker, H., Kammann, C., et al. (2017). Synergistic use of peat and charred material in growing media – An option to reduce the pressure on peatlands? Journal of Environmental Engineering and Landscape Management, 25(2), 160–174. https://doi.org/10.3846/16486897.2017.1284665.
Kim, Y. C., & Anderson, A. J. (2018). Rhizosphere pseudomonads as probiotics improving plant health. Molecular Plant Pathology, 19(10), 2349–2359. https://doi.org/10.1111/mpp.12693.
Kumar, A., Elad, Y., Tsechansky, L., Abrol, V., Lew, B., Offenbach, R., & Graber, E. R. (2018). Biochar potential in intensive cultivation of Capsicum annuum L. (sweet pepper): Crop yield and plant protection. Journal of the Science of Food and Agriculture, 98(2), 495–503. https://doi.org/10.1002/jsfa.8486.
Lugtenberg, B., & Kamilova, F. (2009). Plant-growth-promoting rhizobacteria. Annual Review of Microbiology, 63, 541–556. https://doi.org/10.1146/annurev.micro.62.081307.162918.
Manfredi, S., & Pant, R. (2013). Improving the environmental performance of bio-waste management with life cycle thinking (LCT) and life cycle assessment (LCA). International Journal of Life Cycle Assessment, 18(1), 285–291. https://doi.org/10.1007/s11367-012-0497-5.
Martinez, C., Avis, T. J., Simard, J. N., Labonte, J., Bélanger, R. R., & Tweddell, R. J. (2006). The role of antibiosis in the antagonism of different bacteria towards Helminthosporium solani, the causal agent of potato silver scurf. Phytoprotection, 87(2), 69–76.
Meller Harel, Y., Elad, Y., Rav-David, D., Borenstein, M., Shulchani, R., Lew, B., & Graber, E. R. (2012). Biochar mediates systemic response of strawberry to foliar fungal pathogens. Plant and Soil, 357(1), 245–257. https://doi.org/10.1007/s11104-012-1129-3.
Mulet, M., Bennasar, A., Lalucat, J., & García-Valdés, E. (2009). An rpoD-based PCR procedure for the identification of pseudomonas species and for their detection in environmental samples. Molecular and Cellular Probes, 23(3–4), 140–147.
Mulet, M., Lalucat, J., & García-Valdés, E. (2010). DNA sequence-based analysis of the pseudomonas species. Environmental Microbiology, 12(6), 1513–1530.
Nijhuis, E. H., Pastoor, R., & Postma, J. (2010). Specific detection of Lysobacter enzymogenes (Christensen and Cook 1978) strain 3.1T8 with TaqMan® PCR. Journal of Applied Microbiology, 108(4), 1155–1166.
Noble, R., & Coventry, E. (2005). Suppression of soil-borne plant diseases with composts: A review. Biocontrol Science and Technology, 15(1), 3–20.
Pane, C., Spaccini, R., Piccolo, A., Scala, F., & Bonanomi, G. (2011). Compost amendments enhance peat suppressiveness to Pythium ultimum, Rhizoctonia solani and Sclerotinia minor. Biological Control, 56(2), 115–124. https://doi.org/10.1016/j.biocontrol.2010.10.002.
Parnell, J. J., Berka, R., Young, H. A., Sturino, J. M., Kang, Y., Barnhart, D. M., & DiLeo, M. V. (2016). From the lab to the farm: An industrial perspective of plant beneficial microorganisms. Frontiers in Plant Science, 7(1110), 1–12. https://doi.org/10.3389/fpls.2016.01110.
Postma, J., Montanari, M., & Van Den Boogert, P. H. J. F. (2003). Microbial enrichment to enhance the disease suppressive activity of compost. European Journal of Soil Biology, 39(3), 157–163.
Postma, J., Schilder, M. T., Bloem, J., & Van Leeuwen-Haagsma, W. K. (2008). Soil suppressiveness and functional diversity of the soil microflora in organic farming systems. Soil Biology and Biochemistry, 40, 2394–2406.
Postma, J., Nijhuis, E. H., & Someus, E. (2010). Selection of phosphorus solubilizing bacteria with biocontrol potential for growth in phosphorus rich animal bone charcoal. Applied Soil Ecology, 46(3), 464–469.
Postma, J., Clematis, F., Nijhuis, E. H., & Someus, E. (2013). Efficacy of four phosphate-mobilizing bacteria applied with an animal bone charcoal formulation in controlling Pythium aphanidermatum and Fusarium oxysporum f.sp. radicis lycopersici in tomato. Biological Control, 67(2), 284–291.
Pugliese, M., Liu, B., Gullino, M. L., & Garibaldi, A. (2011). Microbial enrichment of compost with biological control agents to enhance suppressiveness to four soil-borne diseases in greenhouse. Journal of Plant Diseases and Protection, 118(2), 45–50. https://doi.org/10.1007/bf03356380.
Rademaker, J. L. W., Louws, F. J., & De Bruijn, F. J. (1998). Characterization of the diversity of ecologically important microbes by rep-PCR genomic fingerprinting. In A. D. L. Akkermans, J. D. Van Elsas, & F. J. De Bruijn (Eds.), Molecular Microbial Ecology Manual (pp. 1–27). Dordrecht: Kluwer Academic Publishers.
Reid, A., & Greene, S.E. (2012). How microbes can help feed the world. Report on an American Academy of Microbiology Colloquium (pp. 33). Washington DC.
Schnug, E., & Lottermoser, B. G. (2013). Fertilizer-derived uranium and its threat to human health. Environmental Science & Technology, 47, 2433–2434.
Smith, C. J., & Osborn, A. M. (2009). Advantages and limitations of quantitative PCR (Q-PCR)-based approaches in microbial ecology. FEMS Microbiololy Ecology, 67, 6–20.
Someus, E., & Pugliese, M. (2018). Concentrated phosphorus recovery from food grade animal bones. Sustainability, 10(7), 2349.
Termorshuizen, A. J., van Rijn, E., van der Gaag, D. J., Alabouvette, C., Chen, Y., Lagerlof, J., et al. (2006). Suppressiveness of 18 composts against 7 pathosystems: Variability in pathogen response. Soil Biology and Biochemistry, 38(8), 2461–2477.
Trillas, M. I., Casanova, E., Cotxarrera, L., Ordovás, J., Borrero, C., & Avilés, M. (2006). Composts from agricultural waste and the Trichoderma asperellum strain T-34 suppress Rhizoctonia solani in cucumber seedlings. Biological Control, 39(1), 32–38. https://doi.org/10.1016/j.biocontrol.2006.05.007.
United Nations (2017). World population prospects: The 2017 revision, key findings and advance tables. Working Paper No. ESA/P/WP/248: Department of Economic and Social Affairs, Population Division.
Vandecasteele, B., Debode, J., Willekens, K., & Van Delm, T. (2018). Recycling of P and K in circular horticulture through compost application in sustainable growing media for fertigated strawberry cultivation. European Journal of Agronomy, 96, 131–145. https://doi.org/10.1016/j.eja.2017.12.002.
Vestberg, M., Kukkonen, S., Parikka, P., Yu, D., & Romantschuk, M. (2014). Reproducibility of suppression of pythium wilt of cucumber by compost. Agricultural and Food Science, 23(3), 236–245.
Wang, D., Yu, J. M., Pierson Iii, L. S., & Pierson, E. A. (2012). Differential regulation of phenazine biosynthesis by RpeA and RpeB in pseudomonas chlororaphis 30-84. Microbiology (United Kingdom), 158(7), 1745–1757. https://doi.org/10.1099/mic.0.059352-0.
Warren, G. P., Robinson, J. S., & Someus, E. (2009). Dissolution of phosphorus from animal bone char in 12 soils. Nutrient Cycling in Agroecosystems, 84(2), 167–178.
Yamamoto, S., Kasai, H., Arnold, D. L., Jackson, R. W., Vivian, A., & Harayama, S. (2000). Phylogeny of the genus pseudomonas: Intrageneric structure reconstructed from the nucleotide sequences of gyrB and rpoD genes. Microbiology, 146(10), 2385–2394.
Ye, J., Coulouris, G., Zaretskaya, I., Cutcutache, I., Rozen, S., & Madden, T. L. (2012). Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics, 13, 134–134. https://doi.org/10.1186/1471-2105-13-134.
Acknowledgements
The research was financially supported by the 7th Framework of the European Commission, grant agreement no. 289785 (REFERTIL, http://www.refertil.info). We thank Edward Someus and Massimo Pugliese for coordinating and organising the organic products. Compost and biochar products were kindly supplied by Van Iersel, Proficomp, BGU, Pyreg and Terra Humana. We gratefully acknowledge the chemical analyses performed by Wessling Hungary Kft. and technical support at the experiments by Mirjam T. Schilders and Hanneke A.G. Elema.
Funding
This study was funded by 7th Framework of the European Commission (grant agreement no. 289785).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
This article does not contain any study with human participants or animals performed by the authors.
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
ESM 1
(PDF 294 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Postma, J., Nijhuis, E.H. Pseudomonas chlororaphis and organic amendments controlling Pythium infection in tomato. Eur J Plant Pathol 154, 91–107 (2019). https://doi.org/10.1007/s10658-019-01743-w
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-019-01743-w