Abstract
Tomato (Solanum lycopersicum) is a model for climacteric fleshy fruit ripening studies. Tomato ripening is regulated by multiple transcription factors together with the plant hormone ethylene and their downstream effector genes. Transcription Factors APETALA2a (AP2a), NON-RIPENING (NOR) and FRUITFULL (FUL1/TDR4 and FUL2/MBP7) were reported as master regulators controlling tomato fruit ripening. Their proposed functions were derived from studies of the phenotype of spontaneous mutants or RNAi knock-down lines rather than, as it appears now, actual null mutants. To study TF function in tomato fruit ripening in more detail, we used CRISPR/Cas9-mediated mutagenesis to knock out the encoding genes, and phenotypes of these mutants are reported for the first time. While the earlier ripening, orange-ripe phenotype of ap2a mutants was confirmed, the nor null mutant exhibited a much milder phenotype than the spontaneous nor mutant. Additional analyses revealed that the severe phenotype in the spontaneous mutant is caused by a dominant-negative allele. Our approach also provides new insight into the independent and overlapping functions of FUL1 and FUL2. Single and combined null alleles of FUL1 and FUL2 illustrate that these two genes have partially redundant functions in fruit ripening, but also unveil an additional role for FUL2 in early fruit development.
Similar content being viewed by others
Introduction
Tomato (Solanum lycopersicum) produces fleshy fruits, which are climacteric, i.e. displaying a burst in ethylene production during ripening. Its diploid genome, available genome sequence and relative ease of transformation make it the ideal model for studying fleshy fruit development and ripening.
Biochemical and physiological processes during tomato fruit ripening result in changes in texture, colour and flavour. Together with ethylene, transcription factors (TFs) and their downstream effector genes regulate these changes1. RIPENING INHIBITOR (RIN), COLORLESS NON-RIPENING (CNR), TOMATO AGAMOUS-LIKE1 (TAGL1), APETALA2a (AP2a), NON-RIPENING (NOR) and FRUITFULL (FUL1 and FUL2) are major TFs regulating tomato fruit ripening, either by promoting or repressing this process1. RIN, TAGL1, and FRUITFULL1 and 2 are Minichromosome Maintenance (MCM1), AGAMOUS (AG), DEFICIENS (DEF) and Serum Response Element (SRF) (MADS) domain TFs2,3, are highly expressed during the ripening stage and were reported as positive regulators of tomato fruit ripening4,5. MADS domain proteins often function as a dimer or tetramer for regulation6 and interaction between RIN and TAGL1 or FUL was shown by yeast-2-hybrid studies7. Tomato SQUAMOSA promoter-binding protein-like (SPL) transcription factor CNR is also an activator of tomato fruit ripening, involving in ethylene and lycopene biosynthesis. The spontaneous Cnr mutant shows a colourless pericarp with strongly reduced ethylene production8,9.
The tomato transcription factor AP2a is a member of the APETALA2/Ethylene Response Factor (AP2/ERF) family10. Using RNAi Chung et al.11 and Karlova et al.12 showed that the down regulation of AP2a interfered with normal ripening in fruits, including decreased carotenoid production, but increased ethylene production resulting in early onset of fruit ripening and senescence. Thus, AP2a is a negative regulator of ethylene production, but a positive regulator of other ripening aspects such as chlorophyll degradation and carotenoid biosynthesis. A negative feedback loop of AP2a and CNR during ripening was reported, in which AP2a was regulated by RIN, NOR and CNR, while AP2a itself negatively regulates CNR12. AP2a is also a target of post-transcriptional regulation by miR17213. NAC-NOR is a NAM, ATAF1/2 and CUC2 (NAC) domain transcription factor, containing the conserved NAC domain that functions in DNA binding as well as in dimerization with other NAC proteins14,15. There are 101 NAC genes in tomato but, only three (NAC1, NAC4 and NAC-NOR) were shown to be involved in regulation of fruit ripening so far16,17. NAC-NOR appeared to be the most strongly regulating activator based on the completely non-ripening phenotype of its spontaneous mutant18. In the spontaneous nor mutant, a 2 bp deletion in the third exon of NAC-NOR causes a frameshift, resulting in a truncated protein giving a strong non-ripening phenotype19. Tomato MADS domain transcription factors FUL1 and FUL2 are co-orthologs of Arabidopsis FRUITFULL20. FUL2 is expressed in flowers and developing green fruits, and its expression increases during ripening. FUL1 expression is detectable in flowers, but in fruits it is much higher and specific for the ripening stage20. Yeast-2-hybrid protein interaction experiments showed that both could interact with RIN, which is also expressed during ripening, while FUL2 interacts with other MADS domain proteins as well5,7. RNAi experiments showed that FUL1 and FUL2 probably function redundantly in tomato fruit ripening20,21,22. Phenotypes of FUL1/2 RNAi fruits diverged between studies, showing an orange-ripe phenotype with reduced lycopene level and relatively normal ethylene production in one study20, and almost green fruits with strongly reduced ethylene production in another21,22.
In the absence of available spontaneous mutants Virus-induced Gene Silencing (VIGS) of gene expression and RNA interference (RNAi) have often been used for evaluating gene function. Both approaches however, may suffer from incomplete suppression of expression or lack of specificity for the targeted gene. Because RNAi silencing was the most popular tool in the past decades due to the relative ease of use23, functional characterization of the gene of interest may have been imperfect in many cases. The action of Site-Specific Nucleases (SSN) allows targeted mutagenesis by utilizing the imperfect nature of double-strand DNA break (DSB) repair, creating mostly small INDELs which, when located in an open reading frame, can lead to frame-shifts resulting in loss-of-function alleles. Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/ CRISPR-associated protein 9 (Cas9) has rapidly gained popularity as the SSN of choice for mutagenesis due to its high efficiency and relative ease of use. It utilizes guideRNAs (gRNAs), which recognise the target sequence to direct the endonuclease Cas9 to cut there, causing a DSB24. Together with efficient modular cloning strategies such as Golden Gate cloning25 it allows multiple gRNAs targeting more than one gene at the same time with high efficiency26, and CRISPR/Cas9-mutagenesis has been successfully applied in many plant species, such as Arabidopsis, rice, maize and tomato27.
Spontaneous mutants with fruit ripening phenotypes in tomato have been reported for decades and forward genetics studies have identified several of the underlying genes as encoding transcription factors, such as for the rin and nor mutants. These mutants have proven extremely valuable for both fundamental research as well as in applications. Yet, the availability of a larger set of alleles may improve our understanding of TF function further, as well as allow study of specific, true knock-out phenotypes where only RNAi studies were available before. Recently, by using CRISPR/Cas9-mediated mutagenesis Ito et al. illustrated that the rin phenotype is caused by the production of a fusion protein RIN-MC, rather than the mere loss of function of MADS-RIN28. After knocking out RIN in a wild type background, fruit ripening was affected but not blocked as it was in the original rin mutant29, and ripening was partially restored by knocking out RIN-MC in the rin background. There are so far no spontaneous mutants of AP2a, FUL1 or FUL2 reported, and RNAi or VIGS phenotypes may only partially reflect the functions of these genes.
In this study, by using CRISPR/Cas9-mutagenesis we generated genuine knock-out mutants of AP2a, NAC-NOR, FUL1 and FUL2 (as well as the latter two combined) to further study their function in tomato fruit ripening. In this way we confirm the previously found function of AP2a, but demonstrate that the spontaneous nor mutation represents a dominant-negative allele of NAC-NOR because a null allele has a milder phenotype. Moreover, true knock-out mutants of FUL1 and FUL2 allowed to differentiate between shared functions during ripening and a specific FUL2 function in early fruit development.
Results and Discussion
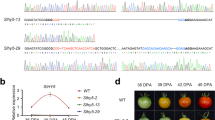
In order to obtain knock-out mutations in the selected transcription factor genes in tomato cv. Moneyberg, we have used binary vectors containing SpCas9 combined with 2 guideRNA-encoding expression cassettes. Two guides were used to target a single gene or, combining two guides, one for FUL1 and FUL2 each, to produce ful1/ful2 double mutants. In all cases, primary transformants were genotyped for targeted mutations, selected transformants were selfed and only homozygous or biallelic mutants were used for phenotyping. The locations of the targeted sites as well as the obtained (-cr) alleles are shown in Fig. 1 and Supplementary Fig. 1. As can be seen in the latter, and since mutant alleles were derived from just one or two distinct guideRNAs, similar detrimental effects on the resulting protein functions were expected, and we selected those which were deemed representative for all similar mutations. Flowers were labelled at anthesis as 0 Days Post Anthesis (DPA) to record the time required to reach the Breaker (Br) stage, and ethylene production of fruits was measured at Breaker and Breaker + 5 days stages.
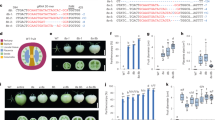
Targets for CRISPR/CAS9 mutagenesis of tomato transcription factor genes, and resulting mutant alleles. (a) Mutations in AP2a. Overview of the AP2a gene and protein changes in knock-out mutants. sgRNA AP2a-t1 and AP2a-t2 located in the first exon were used. (b) Mutations in NAC-NOR. Overview of the NAC-NOR gene and protein changes in the spontaneous nor (nor-s) and CRISPR alleles. sgRNA NOR-t1 and NOR-t6 were designed at the start and middle of the NAC domain for all mutagenesis experiments. (c) Mutations in FUL1. FUL1 gene and protein changes in knock-out alleles. (d) Mutations in FUL2. Overview of the FUL2 gene and protein changes in CRISPR alleles. Regions in orange, pink and red represent the AP2, NAC and MADS domain, respectively. Letters in red indicate spacer sequences and underlined are protospacer adjacent motifs (PAM). The start codon is indicated with red boxes. Numbers represent the location of the nucleotide in the coding sequence. A black diamond shows the single amino acid deletion in ful2-cr2.
ap2a mutants initiate fruit ripening earlier, but do not fully ripen
AP2a (Solyc03g044300) was reported to be a negative regulator of tomato fruit ripening initiation based on RNAi suppression of expression12, but true knock-out mutants were not available so far. The encoded protein of 401 amino acids contains two AP2 domains (amino acids 135 to 201 and 227 to 294), presumably involved in DNA binding30. By using two gRNAs, four alleles ap2a-cr1- 4 were obtained with deletions in the first of 9 exons (Fig. 1a and Supplementary Fig. 1a). We selected two lines with deletions most probably resulting in null alleles. In ap2a-cr1 a 35 bp deletion joining the two gRNA-target sites is predicted to produce a peptide of 27 aa with no AP2 domains, while the 133 bp deletion in allele ap2a-cr2 extends to 67 nucleotides upstream of the start codon, and therefore no AP2a protein is expected to be produced (Fig. 1a). The 133 bp deletion in ap2a-cr2 extends into the 5′ UTR of AP2a, which does not necessarily affect its transcription. This mutation deletes the first start codon, as well as an alternative start codon at amino acid position 12. The next in-frame start codon at amino acid position 204 is located in exon 5 and 3′ of the first of two conserved AP2 domain-encoding regions and therefore even if used, unlikely to result in a functional protein.
When compared to wild type fruits (Fig. 2a) pericarp of the two lines remained orange/brown (Fig. 2b,c) 20 days after Breaker stage and did not become fully red (Supplementary Fig. 2a). Faster ripening was accompanied by earlier senescence: ap2a-cr1 and ap2a-cr2 fruits started to crack before 60 DPA while fruits from other mutants were still intact (Supplementary Fig. 2a). Fruits of the ap2a mutants took only 39 to 41 days to reach Br stage in ap2a-cr1 and ap2a-cr2, respectively, significantly less than in wild type fruits (47 days) (Fig. 3a). This is consistent with the observed 7 days earlier colour change in RNAi fruits11, confirming AP2a’s negative regulatory role in the initiation of tomato fruit ripening. Fruits of both ap2 knock-out mutants produced at least twice the wild type amount of ethylene at Br stage (Fig. 3b).
Differences in developmental and ripening processes of mutants compared to wild type. (a) Time to initiation of ripening (Days Post Anthesis (DPA) to Breaker) of wild type and homozygous mutants. (b) Ethylene production (ppm/g/h) for wild type and homozygous mutants at Br and Br+ 5 d stages and of the spontaneous nor mutant at the equivalent stage. Values of five or six fruits were used. (c) Relative AP2a expression in two ap2a knock-out lines. Error bars represent SE of means. (d) Diameter (cm), (e) Height (cm) and (f) Weight (g) of wild type and homozygous ful2 mutant fruits. Values of eleven fruits for each genotype were used for (d–f). Asterisks show significant differences (P < 0.05).
The AP2 protein in Arabidopsis is known to negatively regulate its own transcription by a feedback regulation31. While RNAi knock-down of expression usually reduces mRNA levels of the target gene with varying efficacy, remaining mRNA can still produce functional protein. Mutants producing no functional proteins, apart from giving a stronger phenotype also allow the exploration of positive or negative feedback autoregulation of the protein on its own expression. This is exemplified by expression analysis of the tomato AP2a mRNA in wild type and ap2a-cr1 and -cr2 fruits. As the mutations result in no functional protein production, mutant fruits display higher levels of AP2a mRNA, indicating that similar to Arabidopsis AP2, tomato AP2a protein negatively regulates its own transcription (Fig. 3c). The much higher ethylene production and faster ripening in ap2a-cr1 and ap2a-cr2 confirm AP2a’s negative role in both ethylene production as well as in initiation of tomato fruit ripening. Its positive regulatory role in ripening is shown by the orange/brown fruit colour resulting from a lack of lycopene production and a defect in chlorophyll degradation, consistent with the RNAi phenotype from Karlova et al.12.
A nor null mutant has a milder phenotype than the spontaneous nor mutant
Ripening defects in the spontaneous nor mutant are likely due to a 2 bp deletion in the third exon of NAC-NOR (Solyc10g006880), leading to a frame shift and a truncated protein of 186 amino acid (aa) versus 355 aa for the intact gene32. This truncation is located after the NAC domain, leaving the possibility that the truncated protein retains its dimerization and DNA-binding capacity. To knock out NOR in cv. Moneyberg we designed two gRNAs for NAC-NOR but found no edits at position t6. Two alleles at position t1 were obtained, nor-cr1 with a 1 bp deletion and nor-cr3 with a 2 bp deletion 5′ of the NAC-domain coding sequence (Fig. 1b and Supplementary Fig. 1b), both resulting in a frameshift and a protein predicted to contain only 17 aa of NAC-NOR and no conserved NAC domain (Fig. 1b).
Homozygous nor-cr1 fruits initiated ripening later than wild type fruits (Fig. 2d) by 3 days on average (time to Br stage, Fig. 3a), but surprisingly progress of ripening was only partially affected, in contrast to being totally blocked as it is in the spontaneous mutant (Fig. 2e). Homozygous nor-cr1 exhibited an orange pericarp at 60 DPA, indicating that lycopene biosynthesis was affected. Colour change after Breaker in nor-cr1 was much slower than in wild type fruits and the pericarp remained orange until 70 DPA and beyond (Supplementary Fig. 2b). Ethylene production in nor-cr1 fruits at both Br and Br+ 5 d stages was significantly lower than in wild type fruits, possibly explaining the delayed initiation of ripening, but clearly higher than in spontaneous nor mutant fruits, where no ethylene production was detectable in the time frame where normally ripening occurs (Fig. 3b).
The alcobaca (alc) mutation, encoding a deleterious V106D substitution in the NAC domain is allelic to nor and displays a weaker effect on ripening33. By CRISPR/Cas9-induced gene replacement Yu et al. replaced thymine to adenine at position 317 of the NAC-NOR coding sequencing, creating an alc allele and confirming the long-shelf life character of alc34. Three Penjar accessions contain the alc allele, while a fourth contains an early frame-shift and a truncated protein of 6 aa, which is similar to the nor-cr1 allele described here. Our nor-cr1 fruits displayed delayed ripening and an orange-ripe phenotype similar to that of alc fruits. As the truncated protein in nor-cr1 contains no NAC domain this likely makes nor-cr1 a true null allele. A transcriptional activation region located in the C-terminal region of NAC proteins is essential for activating transcription35. Candidates for interaction with NAC-NOR would be NAC-NOR itself forming homodimers or the two other tomato NAC proteins involved in ripening, NAC1 and NAC4, and such an interaction was demonstrated for NAC416. The decreased ethylene production in nor-cr1 confirms that NAC-NOR, as a master regulator, is located upstream of ethylene biosynthesis and has a positive regulatory role.
The spontaneous nor mutation produces a dominant-negative protein
Ripening in the spontaneous nor mutant, whose allele we shall from here on call nor-s for convenience, is totally blocked but only partially so in nor-cr1 and alc fruits. Therefore we hypothesize that the non-ripening phenotype in nor-s is caused by the truncated protein functioning in a dominant-negative manner, where the protein product is able to interact with other NAC proteins and to bind DNA without transcriptionally activating its targets. This is reminiscent of the NAC TF SND1 in poplar, where an alternative splice variant retains the last intron and due to a premature stop forms a protein without an activation domain but with an almost intact NAC domain. This protein acts as a dominant-negative repressor of its downstream targets as well as of its own and family members’ expression36. This is also reminiscent of the situation with the mads- rin allele blocking ripening, although there the newly formed RIN-MC fusion protein has a novel combination of expression and transcriptional activation not seen in a rin knock-out line28,37. To further study this we introduced mutations 5′ of the location of the spontaneous mutation in NOR in both the spontaneous nor mutant as well as in the wild type Ailsa Craig background. Two alleles, nor-scr1 and nor-scr2 with a 1 bp deletion and insertion, respectively, were obtained in the spontaneous nor-s mutant background, and one allele (nor-cr2) with the same 1 bp insertion as nor-scr2 was obtained in wild type Ailsa Craig (Fig. 1b). Homozygous nor-cr2 caused an orange-ripe pericarp in Ailsa Craig fruits, and ripening in homozygous nor-scr2 and biallelic nor-scr1/nor-scr2 mutants was similar to this (Fig. 4). Therefore we can conclude that the frameshift upstream of the spontaneous mutation precludes translation of the NAC domain and negates the dominant-negative function of the spontaneous mutant, which retained an intact NAC domain.
A classical dominant- negative TF would still interact with the same regulatory DNA elements or form the same dimers as the wild type protein, but the activity of the dimer in a heterozygote would only be 25% compared to homozygous wild type thus giving a phenotype more similar to that of the homozygous mutant38. Heterozygous nor-s mutants in cv. Rutgers have an intermediate phenotype, both in timing of ripening, carotene production, as well as in ethylene production during ripening39. This indicates that nor-s’ negative effect is dose-dependent. This suggests that nor-s is a so-called trans-acting dominant-negative allele having interlocus interactions, rather than the classical intralocus interaction38. The tomato NAC1 and NAC4 TFs (see previous section) would be obvious candidates for such intralocus interactions.
FUL1 and FUL2 have overlapping functions during fruit ripening, but FUL2 has an additional role in fruit development
Since tomato FUL1 (Solyc06g069430) and FUL2 (Solyc03g114830) are close paralogs, it was particularly challenging to achieve RNAi-mediated knock-down of expression in a specific as well as effective manner20. Use of a less specific RNAi construct might have led to knock-down of multiple homologous genes. Both studies reporting knock-down of tomato FUL genes achieved knock-down of both concomitantly21,40, but only relatively weak and not completely specific knock-down of each gene individually in another study20. These results suggest that FUL1 and FUL2 were functioning at least partially redundant in tomato fruit ripening, but were inconclusive about the relative roles of the two genes. We therefore generated both ful1 and ful2 single mutants as well as double mutants using CRISPR/Cas9. We obtained multiple knock-out alleles in both FUL1 and FUL2, alone or in combination. Double mutants were obtained with constructs containing sgRNAs for both genes. A ful1 single mutant line containing the ful1-cr1 allele with a 91 bp deletion, presumably caused by microhomology-directed repair of the double strand break, which completely deleted the second exon, produces a truncated protein of 62 aa consisting of the MADS domain only (Fig. 1c). The 1 bp deletion in mutant allele ful1-cr2 leads to a truncation at amino acid position 105 (Fig. 1c). Both alleles disable FUL1 function. In the ful2 single mutant, the ful2-cr1 allele has a 1 bp insertion in the middle of the MADS domain-encoding region resulting in a truncated protein with only 29 aa (Fig. 1d). A 3 bp deletion in FUL2 in the ful2-cr2 allele allows production of the entire protein minus one amino acid, arginine 25 in the middle of the extremely conserved MADS domain. This mutation is very likely to be deleterious to protein function, as further supported by analysis in the Provean protein website (Provean score: −12.037)41. Other obtained alleles are shown in Supplementary Fig. 1c,d. We also checked the sequences of the corresponding parts in the non-target paralog for FUL1 and FUL2 (Supplementary Fig. 1c,d) reciprocally but found no mutations there, demonstrating the high specificity of sgRNAs.
There were no apparent differences in final overall fruit colour between ful1-cr1, ful2-cr1 and wild type fruits at 55 DPA (Fig. 2a,f,g). However, in both double mutants (Fig. 2h,i) the pericarp stayed orange until 60 DPA (Supplementary Fig. 2a) and did not reach a red ripe colour as in the wild type fruits (Fig. 2a and Supplementary Fig. 2a), which is similar to the phenotype observed for the RNAi FUL1/FUL2 silenced lines in the study of Bemer et al.20.
Interestingly, we observed a phenotype in ful2-cr mutants that had not been described before in the RNAi knock-down lines. In early stages of fruit development, superficial stripes lighter than the surrounding pericarp were visible at the bottom of homozygous ful2-cr1 fruits, but not in homozygous ful1-cr1 fruits, which became less distinguishable as fruits ripened (Figs 2g, 5a and Supplementary Fig. 2c). Significantly lighter-pigmented stripes were also found at the bottom of all fruits in the two double mutants ful1-cr2/ful2-cr2 and ful1-cr2/ful2-cr3, which only changed from white to yellow around the time when wild type fruits are fully ripe (Figs 2h,i and 5a). These stripes were not only visible superficially, but also in the mesocarp of sections of both homozygous ful1/ful2 double mutants, making the pericarp at the bottom region and septum much lighter coloured than the rest of the fruit (Fig. 5b). Moreover, vertical, possibly suberized cracks in the surface of all ful2 mutant lines, including double mutant lines, were visible from the early stages of fruit development (Figs 2g–i and 5c,d). Additionally, the columella and placenta of ful2 and ful1/ful2 fruits remained white when fruits were fully ripe (Fig. 5b,d). Besides, we noticed that fruits of all the mutants with a ful2 null allele were more than 10 mm smaller than wild type fruits, both in diameter and in height (Fig. 3d,e) and almost half the weight of wild type fruits (Fig. 3f). Also there was a significant effect of ful2 on time to onset of ripening (Br stage). It took 46 days in ful1-cr1 and only 43 days from anthesis to Breaker in ful2-cr1 fruits, significantly less than in wild type fruits (Fig. 3a and Supplementary Fig. 2a). The time to ripening was also significantly shorter than in wild type fruits in both double mutant lines (Fig. 3a), but the pericarp stayed orange until 60 DPA and did not reach a red ripe colour as in the wild type fruits. We speculate that the smaller fruit size of ful2 mutants may have a causal relation to the earlier ripening phenotype, but this will require more study.
Details of fruit phenotypes and differences in ful2 mutants compared to wild type. (a) Bottom view of ful2 single and double mutants. Light coloured stripes only at the bottom of ful2 mutant fruits appear from early green fruit stage. (b) Longitudinally sliced fruits of ful2 null mutants with light coloured-pericarp at the bottom of fruits. (c) Details of cracks in ful2 mutants. Cracks in all ful2 mutants at unripe (top), including details (middle) and ripe (bottom) stage. Black arrows indicate cracks in unripe fruits. (d) Latitudinally sliced fruits of ful2 mutants. Boxed regions are enlarged in the lower row. Scale bar, 1 cm.
We also measured ethylene production in all mutants, and found significantly lower production in the ful1-cr1 single mutants and the double mutants, but not so in the ful2-cr1 mutants (Fig. 3b). The strong reduction in the ful1-cr mutant indicates that FUL1 is an important regulator of ethylene biosynthesis in the ripening phase, although its reduction in the single mutant has little effect on visible ripening aspects. However, FUL2 also contributes, because ethylene production is even more reduced in the ful1-cr2/ful2-cr3 double mutants. Compared to wild type fruits, ethylene production decreased to 32% and 17% in two double mutant lines at Breaker stage, and to only 15% and 5% at Br+ 5 d (Fig. 3b). Thus, although FUL1 and FUL2 were reported to act redundantly during fruit ripening, ful2 mutants affect unique aspects of fruit development, both time to ripening and as well as fruit skin integrity, which are similarly affected in double mutants. By contrast, ful1-cr1 showed a very mild difference from WT without cracks or stripes in the pericarp, nor a difference in time to ripening, but a stronger effect on ethylene production. This is consistent with the strong increase in FUL1 expression during ripening, while FUL2 is expressed in developing green fruits as well. Some of the differences in the functions of FUL1 and FUL2 that have become apparent in this study may also reflect tissue-specific and temporal differences in expression. According to the Tomato Expression Atlas (TEA)42 FUL2 expression is especially high in internal fruit tissues (columella, placenta, locule tissues) while FUL1 expression, during ripening, is more evenly distributed over pericarp and internal tissues (Supplementary Fig. 3). FUL2 expression however is higher than FUL1 expression in all fruit tissues up to the mature green stage.
Phenotypes of ful1 or ful2 null alleles are reported for the first time in our study and although their combined mutations had a severe ripening phenotype, they are very different from FUL1/FUL2 double knock-down lines reported by Shima et al. or Wang et al., in which they showed a totally blocked ripening phenotype21 or bright yellow fruits40 when silencing both FUL1 and FUL2. The orange pericarp of ful1/ful2 true knock-out mutants is similar to what Bemer et al.20 presented in their RNAi experiment, while single mutants exhibited normal red-ripe pericarp, confirming their redundant function in fruit ripening. Light coloured stripes at the bottom region and vertical (possibly suberized) cracks in the surface of all lines containing homozygous ful2 alleles have never been reported before. They are visible from the green fruit stage, illustrating additional roles of FUL2 in early fruit development and in carotenoid biosynthesis. Fujisawa et al. showed that the promoter of PHYTOENE SYNTHASE 1 (PSY1) was a direct target of RIN and FUL1, but not of FUL222, suggesting that another mechanism is responsible for FUL2 regulating fruit pigmentation. Possibly FUL2 is involved in chloroplast formation, stability, or function during early fruit development, which would account for the lighter green areas. The cracks in the surface of all ful2 (both single and double mutant) lines are a unique phenotype of ful2 mutants that is not shared with ful1 single mutants. The aforementioned RNAi studies of combined FUL1/FUL2-function all reported thinner-cuticles, leading to faster water loss in ripe fruits20,40. The observed cracks in the ful2 CRISPR mutants indicate that this may well represent a specific FUL2 function in cutin formation or epidermal development in tomato fruit.
The additive effect in ethylene production among single and double mutants proves the partially redundant functions of FUL1 and FUL2 in ethylene biosynthesis and fruit ripening. In contrast to the unchanged ethylene production in knock-down lines shown by Bemer et al.20, the strongly decreased ethylene level in ful1/2 double knock-out mutants in our study illustrates that FUL1 and FUL2 regulate tomato fruit ripening via ethylene biosynthesis, consistent with other studies21,40. These discrepancies between studies are possibly caused by the use of different genotypes (MicroTom, Ailsa Craig or Moneyberg), and by the limited downregulation in the RNAi lines previously analysed12. Here it is shown that only one third or less of wild type ethylene production in ful1/ful2 double mutants could still support some ripening progression, as it does in the nor-cr mutants indicating that the initiation of ripening may only require a limited amount of ethylene, and once it reaches a threshold, ripening starts even if not progressing to its full extent.
In conclusion, we have demonstrated the utility of CRISPR/Cas9-mutagenesis in tomato for reassessing transcription factor gene functions. Some phenotypes closely resembled those that were previously reported, but in addition allow the study of regulatory features such as an auto feedback regulation of transcription (ap2a) or complementation by retransformation. For others, studying alternative alleles gives more insight into gene function by revealing a distinction between null and dominant-negative alleles (nor-s), respectively. Finally, gene-specific mutations allow the separation of functions (or demonstrate redundancy) of pairs of very similar paralogs (ful1 and ful2), which are difficult to separate by older methods such as RNAi or VIGS.
Materials and Methods
Plant materials and growing conditions
Tomato cv. Moneyberg, Ailsa Craig (AC) and the AC nor mutant (the latter two obtained from the Tomato Genetics Resource Centre, TGRC) were used for the Agrobacterium tumefaciens-mediated transformation experiments43. Tissue culture was done in a growth chamber with 16 h light and 8 h dark at 25 °C. Larger plants before flowering were moved to the greenhouse facilities of Unifarm, Wageningen University & Research and grown and phenotyped under standard greenhouse conditions.
gRNA design and mutagenesis constructs
Online programs CRISPR-P 1.0 (http://crispr.hzau.edu.cn/CRISPR/)44 and CRISPOR (http://crispor.tefor.net/crispor.py)45 were used for designing gRNAs and for excluding off-targets.
The MoClo Toolkit46 was used to assemble constructs with gRNAs targeting each gene and the Golden Gate cloning strategy was used as described earlier to assemble binary vectors for tomato mutagenesis47. Briefly, each gRNA fused to the synthetic Arabidopsis U6 promoter as AtU6p::gRNA was ligated in a Level 1 vector. Level 1 constructs pICH47732-NOSpro::NPTII::OCST, pICH47742-35S::Cas9::NOST, pICH47732-gRNA1, pICH47742-gRNA2 and the linker pICH41780 were cut/ligated into the Level 2 vector pICSL4723 as described48. All primers used for amplifying gRNAs with backbones are listed in Supplementary Table 1.
Transgenic plant genotyping
Genomic DNA from young leaves was isolated using the CTAB method49. PCRs for Cas9 and NPT2 were performed for all regenerated plants and only from the Cas9/NPT2 positive plants target regions were sequenced. Heterozygous and biallelic mutants were selfed and T1 seedlings were screened for the absence of Cas9 and the presences of homozygous mutations. Homozygous mutants without Cas9 after segregation were used for further study. All primers used for genotyping are listed in Supplementary Table 1.
Fruit development phenotyping
Two plants per genotype were used for phenotyping. Tomato flowers of all the mutant lines and WT were vibrated and labelled at the day when they were first fully open as 0 Days Post Anthesis (DPA). Data from at least fifteen flowers/fruits per genotype was used for calculating the time taken to reach Breaker stage. Fruits at 35, 40, 45, 50, 55 and 60 DPA were collected for photography. Ten or eleven fruits at 60 DPA were collected for size and weight measurement. Normal distribution of data was confirmed with the R package version 3.5.0 and ANOVA was used to test for the significance of differences between individual mutants and wild type, respectively.
Ethylene measurements
Tomato fruits at Breaker stage and Breaker + 5 d were harvested. After being in open air for 30 min at room temperature fruits were placed in sealed jars for 3 h. Ethylene concentration was measured when immediately after sealing and after 3 h by injecting 1.5 mL gas to a Focus GC gas chromatograph. Ethylene production was calculated as ppm per gram of fruit per hour (ppm/g/h). Values of five or six fruits per genotype were used for analysis. Since ethylene data did not display a normal distribution, generalized linear model regression with a quasibinomial model was used to determine the significance of differences between genotypes.
Real-time PCR gene expression analysis
RNA was isolated from pericarp of fruits at Br+ 5 d stage by using the InviTrap Spin Plant RNA kit (Stratec) and cDNA was synthesized by the iScript cDNA synthesis kit (Bio-Rad). Two replicates were analysed per plant containing one fruit each from two plants per line. Primers used for qRT-PCR are listed in Supplementary Table 1. iQ SYBR Green Supermix (Bio-Rad) and iCycler iQ5 system (Bio-Rad) were used for quantitative RT PCR. Actin was used as a reference and relative expression changes of AP2a were calculated according to 2−ΔΔCt method as described50. Student’s t-test was performed to detect significant differences.
Data Availability
All data in this study are available in the article and supplementary files, or from the corresponding author upon request. Sequence data can be found in the Sol Genomics Network with the following accession numbers: AP2a (Solyc03g044300 (https://solgenomics.net/locus/17739/view)), NAC-NOR (Solyc10g006880 (https://solgenomics.net/locus/1031/view)), FUL1 (Solyc06g069430 (https://solgenomics.net/locus/581/view)) and FUL2 (Solyc03g114830 (https://solgenomics.net/locus/582/view)).
References
Karlova, R. et al. Transcriptional control of fleshy fruit development and ripening. J. Exp. Bot. 65, 4527–4541 (2014).
Robinson, R. W. & Tomes, M. L. Ripening inhibitor: a gene with multiple effect on ripening. Rep Tomato Genet Coop 18, 36–37 (1968).
Busi, M. V. et al. MADS-box genes expressed during tomato seed and fruit development. Plant Mol. Biol. 52, 801–815 (2003).
Vrebalov, J. et al. Fleshy fruit expansion and ripening are regulated by the tomato SHATTERPROOF gene TAGL1. Plant Cell 21, 3041–3062 (2009).
Shima, Y. et al. Tomato FRUITFULL homologues act in fruit ripening via forming MADS-box transcription factor complexes with RIN. Plant Mol. Biol. 82, 427–438 (2013).
Smaczniak, C., Immink, R. G. H., Angenent, G. C. & Kaufmann, K. Developmental and evolutionary diversity of plant MADS-domain factors: insights from recent studies. Development 139, 3081–3098 (2012).
Leseberg, C. H. et al. Interaction study of MADS-domain proteins in tomato. J. Exp. Bot. 59, 2253–2265 (2008).
Thompson, A. J. et al. Molecular and genetic characterization of a novel pleiotropic tomato-pipening mutant. Plant Physiol. 120, 383–390 (1999).
Manning, K. et al. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat. Genet. 38, 948–952 (2006).
Kim, S., Soltis, P. S., Wall, K. & Soltis, D. E. Phylogeny and domain evolution in the APETALA2-like gene family. Mol. Biol. Evol. 23, 107–120 (2006).
Chung, M.-Y. et al. A tomato (Solanum lycopersicum) APETALA2/ERF gene, SlAP2a, is a negative regulator of fruit ripening. Plant J. 64, 936–947 (2010).
Karlova, R. et al. Transcriptome and metabolite profiling show that APETALA2a is a major regulator of tomato fruit ripening. Plant Cell 23, 923–941 (2011).
Karlova, R. et al. Identification of microRNA targets in tomato fruit development using high-throughput sequencing and degradome analysis. J. Exp. Bot. 64, 1863–1878 (2013).
Olsen, A. N., Ernst, H. A., Leggio, L., Lo & Skriver, K. NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci. 10, 79–87 (2005).
Xie, Q., Frugis, G., Colgan, D. & Chua, N. H. Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 14, 3024–3036 (2000).
Zhu, M. et al. A new tomato NAC (NAM ATAF1/2/CUC2) transcription factor, SlNAC4, functions as a positive regulator of fruit ripening and carotenoid accumulation. Plant Cell Physiol. 55, 119–135 (2014).
Meng, C. et al. Suppression of tomato SlNAC1 transcription factor delays fruit ripening. J. Plant Physiol. 193, 88–96 (2016).
Tigchelaar, E., Tomes, M. & Kerr, E. B. R. A new ripening mutant, non-ripening (nor). Rep. Tomato Genet. Coop. 23, 33–34 (1973).
Giovannoni, J., Tanskley, S., Vrebalov, J. & Noensie, F. NOR gene compositions and methods for use thereof. Patent US 6,762,347 B1 (2004).
Bemer, M. et al. The tomato FRUITFULL homologs TDR4/FUL1 and MBP7/FUL2 regulate ethylene-independent aspects of fruit ripening. Plant Cell 24, 4437–4451 (2012).
Shima, Y. et al. Tomato FRUITFULL homologs regulate fruit ripening via ethylene biosynthesis. Biosci. Biotechnol. Biochem. 78, 231–237 (2014).
Fujisawa, M. et al. Transcriptional regulation of fruit ripening by tomato FRUITFULL homologs and associated MADS box proteins. Plant Cell 26, 89–101 (2014).
Saurabh, S., Vidyarthi, A. S. & Prasad, D. RNA interference: concept to reality in crop improvement. Planta 239, 543–564 (2014).
Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–21 (2012).
Brooks, C., Nekrasov, V., Lippman, Z. B. & Van Eck, J. Efficient gene editing in tomato in the first generation using the Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR-associated9 system. Plant Physiol. 166, 1292–1297 (2014).
Li, J. F. et al. Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 31, 688–691 (2013).
Ma, X., Zhu, Q., Chen, Y. & Liu, Y. G. CRISPR/Cas9 platforms for genome editing in plants: developments and applications. Molecular Plant 9, 961–974 (2016).
Ito, Y. et al. Re-evaluation of the rin mutation and the role of RIN in the induction of tomato ripening. Nat. Plants 3, 866–874 (2017).
Ito, Y., Nishizawa-Yokoi, A., Endo, M., Mikami, M. & Toki, S. CRISPR/Cas9-mediated mutagenesis of the RIN locus that regulates tomato fruit ripening. Biochem. Biophys. Res. Commun. 467, 76–82 (2015).
Jofuku, K. D. Control of Arabidopsis flower and seed development by the homeotic gene. APETALA2. Plant Cell 6, 1211–1225 (1994).
Schwab, R. et al. Specific effects of microRNAs on the plant transcriptome. Dev. Cell 8, 517–527 (2005).
Kumar, R., Tamboli, V., Sharma, R. & Sreelakshmi, Y. NAC-NOR mutations in tomato Penjar accessions attenuate multiple metabolic processes and prolong the fruit shelf life. Food Chem. 259, 234–244 (2018).
Casals, J. et al. Genetic basis of long shelf life and variability into Penjar tomato. Genet. Resour. Crop Evol. 59, 219–229 (2012).
Yu, Q. et al. CRISPR/Cas9-induced targeted mutagenesis and gene replacement to generate long-shelf life yomato lines. Sci. Rep. 7, 11874 (2017).
Puranik, S., Bahadur, R. P., Srivastava, P. S. & Prasad, M. Molecular cloning and characterization of a membrane associated NAC family gene, SiNAC from foxtail millet [Setaria italica (L.) P. Beauv.]. Mol. Biotechnol. 49, 138–150 (2011).
Li, Q. et al. Splice variant of the SND1 transcription factor is a dominant negative of SND1 members and their regulation in Populus trichocarpa. Proc. Natl. Acad. Sci. 109, 14699–14704 (2012).
Li, S. et al. The RIN-MC fusion of MADS-box transcription factors has transcriptional activity. Plant Physiol. 176, 891–909 (2017).
Veitia, R. A. Exploring the molecular etiology of dominant-negative mutations. Plant Cell 19, 3843–51 (2007).
Tigchelaar, E., McGlasson, W. & Franklin, M. Natural and ethephon-stimulated ripening of F1 hybrids of the Ripening Inhibitor (rin) and Non-ripening (nor) mutants of tomato (Lycopevsicon esculentum Mill.). Aust. J. Plant Physiol 5, 449–456 (1978).
Wang, S. et al. Members of the tomato FRUITFULL MADS-box family regulate style abscission and fruit ripening. J. Exp. Bot. 65, 3005–3014 (2014).
Choi, Y., Sims, G. E., Murphy, S., Miller, J. R. & Chan, A. P. Predicting the functional effect of amino acid substitutions and indels. PLoS One 7 (2012).
Shinozaki, Y. et al. High-resolution spatiotemporal transcriptome mapping of tomato fruit development and ripening. Nat. Commun. 9, 364 (2018).
van Roekel, J. S. C., Damm, B., Melchers, L. S. & Hoekema, A. Factors influencing transformation frequency of tomato (Lycopersicon esculentum). Plant Cell Rep. 12, 644–647 (1993).
Lei, Y. et al. CRISPR-P: a web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol. Plant 7, 1494–1496 (2014).
Haeussler, M. et al. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 17, 148 (2016).
Weber, E., Engler, C., Gruetzner, R., Werner, S. & Marillonnet, S. A modular cloning system for standardized assembly of multigene constructs. PLoS One 6 (2011).
Engler, C., Kandzia, R. & Marillonnet, S. A one pot, one step, precision cloning method with high throughput capability. PLoS One 3 (2008).
Werner, S., Engler, C., Weber, E., Gruetzner, R. & Marillonnet, S. Fast track assembly of multigene constructs using golden gate cloning and the MoClo system. Bioeng. Bugs 3, 38–43 (2012).
Porebski, S., Bailey, L. G. & Baum, B. R. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol. Biol. Report. 15, 8–15 (1997).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 (2001).
Acknowledgements
We thank Hani Surya Wijaya and Arne Beukema for technical assistance. Marian Bemer, Vera Veltkamp, and Ellen Slaman are acknowledged for carefully reading and commenting on the manuscript, Marian Bemer and Arjen van de Peppel for assistance with ethylene measurements, and Vera Veltkamp for assistance with statistics in R. This work was supported by a fellowship from the China Scholarship Council (CSC) to R.W., and CAPES/NUFFIC project grant 033/12 to E.C.d.R.T. and A.P.M.
Author information
Authors and Affiliations
Contributions
R.W. and R.A.d.M. designed research; R.W., E.C.d.R.T. and M.L. performed research; R.W., E.C.d.R.T., M.L. and R.A.d.M. analysed data; R.W., E.C.d.R.T., A.P.M., G.C.A. and R.A.d.M. wrote the paper.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, R., Tavano, E.C.d.R., Lammers, M. et al. Re-evaluation of transcription factor function in tomato fruit development and ripening with CRISPR/Cas9-mutagenesis. Sci Rep 9, 1696 (2019). https://doi.org/10.1038/s41598-018-38170-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-38170-6
This article is cited by
-
A tomato NAC transcription factor, SlNAP1, directly regulates gibberellin-dependent fruit ripening
Cellular & Molecular Biology Letters (2024)
-
Transcriptional regulation of tomato fruit ripening
Physiology and Molecular Biology of Plants (2024)
-
Applications of CRISPR/Cas genome editing in economically important fruit crops: recent advances and future directions
Molecular Horticulture (2023)
-
Tailoring crops with superior product quality through genome editing: an update
Planta (2023)
-
Plant tissue culture-mediated biotechnological approaches in Lycium barbarum L. (Red goji or wolfberry)
Horticulture, Environment, and Biotechnology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.