- 1Section Plant Physiology, University of Amsterdam, Amsterdam, Netherlands
- 2Section Plant Cell Biology, Swammerdam Institute for Life Sciences (SILS), University of Amsterdam, Amsterdam, Netherlands
- 3Laboratory of Plant Physiology, Wageningen University and Research, Wageningen, Netherlands
- 4Instituto de Investigaciones Biológicas (IIB-CONICET-UNMdP), Universidad Nacional de Mar del Plata, Mar del Plata, Argentina
Phospholipase C (PLC) has been suggested to play important roles in plant stress and development. To increase our understanding of PLC signaling in plants, we have started to analyze knock-out (KO), knock-down (KD) and overexpression mutants of Arabidopsis thaliana, which contains nine PLCs. Earlier, we characterized PLC2, PLC3 and PLC5. Here, the role of PLC7 is functionally addressed. Promoter-GUS analyses revealed that PLC7 is specifically expressed in the phloem of roots, leaves and flowers, and is also present in trichomes and hydathodes. Two T-DNA insertion mutants were obtained, i.e., plc7-3 being a KO- and plc7-4 a KD line. In contrast to earlier characterized phloem-expressed PLC mutants, i.e., plc3 and plc5, no defects in primary- or lateral root development were found for plc7 mutants. Like plc3 mutants, they were less sensitive to ABA during stomatal closure. Double-knockout plc3 plc7 lines were lethal, but plc5 plc7 (plc5/7) double mutants were viable, and revealed several new phenotypes, not observed earlier in the single mutants. These include a defect in seed mucilage, enhanced leaf serration, and an increased tolerance to drought. Overexpression of PLC7 enhanced drought tolerance too, similar to what was earlier found for PLC3-and PLC5 overexpression. In vivo 32Pi-labeling of seedlings and treatment with sorbitol to mimic drought stress, revealed stronger PIP2 responses in both drought-tolerant plc5/7 and PLC7-OE mutants. Together, these results show novel functions for PLC in plant stress and development. Potential molecular mechanisms are discussed.
Introduction
In animals, phospholipase C (PLC) plays a key role in the perception and transmission of extracellular signals into cells. Activated by extracellular receptors, PLC hydrolyses the membrane phospholipid, phosphatidylinositol 4,5-bisphosphate (PIP2) into two second messengers: inositol 1,4,5-trisphosphate (IP3) and diacylglycerol (DAG). While IP3 triggers the release of Ca2+ from the ER via a ligand-gated Ca2+ channel, the DAG remains in the plasma membrane where it recruitsand activates members of the protein kinase C (PKC) family and stimulates TRP- (Transient Receptor Potential-) channels. Subsequent increases in Ca2+ and phosphorylation status affects various protein targets that regulate multiple processes within and between cells (Irvine, 2006; Michell, 2008; Balla, 2013).
In plants, the PLC-signaling pathway is still enigmatic. Plants lack the primary targets for both IP3 and DAG, and have limited amounts of PIP2 in their membranes (van Leeuwen et al., 2007; Munnik and Nielsen, 2011; Munnik and Zarza, 2013; Munnik, 2014; Heilmann, 2016a,b; Gerth et al., 2017; Noack and Jaillais, 2017). Likely, they use phosphatidylinositol 4-phosphate (PIP) as an additional substrate and phosphorylate the resulting inositol phosphates and DAG into inositol polyphosphates (IPPs; e.g., IP6) and phosphatidic acid (PA), respectively, which can function as second messengers (Munnik, 2001; Gillaspy, 2013; Munnik, 2014; Heilmann, 2016a,b; Hou et al., 2016; Yao and Xue, 2018). IP6 levels increase upon ABA and release Ca2+ in guard cells to induce stomatal closure (Lemtiri-Chlieh et al., 2000, 2003). PA has also been implicated to play a role in ABA signaling, e.g., inhibiting ABI1 (Mishra et al., 2006), activating SnRK2 (Testerink et al., 2004; McLoughlin et al., 2012; Julkowska et al., 2015), and regulating ion channels (Camoni et al., 2012; McLoughlin and Testerink, 2013; Hite et al., 2014). Meanwhile, various other functions for IP6 and additional IPPs have been emerging that could be signaling downstream of PLC, including the pyro-phosphorylated IP7 and IP8 (Laha et al., 2015, 2016). In yeast and mammalian cells, IPP molecules play important roles in various nuclear processes, including gene transcription, chromatin remodeling, mRNA export and DNA repair, involving a wide range of cellular processes, such as osmoregulation, phosphate homeostasis, vesicular trafficking, apoptosis, cell cycle regulation, and ribosome synthesis (Monserrate and York, 2010; Thota and Bhandari, 2015; Williams et al., 2015). In plants, IP6 binds the auxin receptor, TIR1 (Tan et al., 2007), which is proposed to functionally regulate the SCFTIR1 ubiquitin-ligase complex to control downstream auxin mediated-gene expression (Leyser, 2018). Similarly, COI1, the receptor for jasmonate signaling binds IP5 (Sheard et al., 2010) or the pyrophosphorylated form of IP5, i.e., PP-IP5 (= IP7) (Laha et al., 2015, 2016), with functional significance for plant immunity (Mosblech et al., 2008, 2011; Murphy et al., 2008). GLE1, an mRNA export factor, has been identified as an IP6 target in Arabidopsis Pi homeostasis (Lee et al., 2015). SPX domain-containing proteins bind IPPs, including IP6, and many of these proteins are involved in Pi signaling (Kuo et al., 2014, 2018; Puga et al., 2014; Wild et al., 2016). For PA, several plant targets have been identified over the years, including protein kinases, proteins phosphatases, small G-proteins, RBOH (NADPH oxidase), GAPDH, ion channels and actin-binding proteins and PA has been implicated to regulate many cellular processes, including vesicular trafficking, cytoskeleton dynamics, and ion-channels (Munnik, 2001; Wang et al., 2006; Li et al., 2009, 2012; Pleskot et al., 2010, 2017; Testerink and Munnik, 2011; Thomas and Staiger, 2014; Hou et al., 2016; Ufer et al., 2017; Pokotylo et al., 2018; Yao and Xue, 2018). PA is not only generated via PLC and DAG kinase (DGK); it can also be formed via other DAG-generating enzymes, like non-specific PLCs (NPC), or directly, through phospholipase D (PLD) hydrolysis of structural phospholipids (Arisz et al., 2009; Munnik and Testerink, 2009; Pokotylo et al., 2013, 2018; Hou et al., 2016; Yao and Xue, 2018).
How, when, where and whether PLC signaling is involved in generating PA and IPPs is still largely unknown. Hence, tools to genetically manipulate PLC levels would be helpful to functionally address this. As such, silencing of PLC has revealed its importance in plant defense in tomato and Arabidopsis (Vossen et al., 2010; D’Ambrosio et al., 2017), in cytokinins- and gravity signaling in Physcomitrella (Repp et al., 2004), and in ABA signaling and stomatal control in tobacco and Arabidopsis (Sanchez and Chua, 2001; Hunt et al., 2003; Mills et al., 2004). In petunia and tobacco, PLC has been shown to regulate the tip growth of pollen tubes (Dowd et al., 2006; Helling et al., 2006). Arabidopsis T-DNA insertion mutants for PLC3, PLC5 and PLC9 have revealed roles for PLC in seed germination, primary- and lateral root development, ABA signaling and heat stress tolerance (Zheng et al., 2012; Gao et al., 2014; Zhang et al., 2018a,b), while PLC2-insertion mutants showed defects in female gametogenesis and embryo development (Li et al., 2015; Di Fino et al., 2017). Overexpression of PLC has been shown to increase the drought tolerance of maize, canola, tobacco and Arabidopsis (Wang et al., 2008; Georges et al., 2009; Tripathy et al., 2011; Zhang et al., 2018a,b). While it is still not clear how PLC exactly achieves all this, it is important that such molecular tools become available for further physiological- and biochemical analyses.
The Arabidopsis genome encodes 9 PLC genes, which are subdivided into four clades (Hunt et al., 2004; Tasma et al., 2008; Munnik, 2014; Pokotylo et al., 2014). Earlier, we found that knock-out (KO) mutants of PLC3 and a knock-down (KD) mutant of PLC5, exhibited small defects in root development (Zhang et al., 2018a,b). Interestingly, plc3plc5-double mutants did not intensify the phenotype, even though they belong to different clades, indicating that another PLC could be involved. Since PLC3 and PLC5 were both specifically expressed in phloem-companion cells and revealed a “segmented” root-expression pattern from which lateral roots emerge, we searched for additional Arabidopsis PLCs that are phloem-specific and might explain the lack of additional effects on the double mutant. This resulted in the identification of PLC7, which again belongs to another clade. Here, the functional analysis of this PLC is described, revealing novel phenotypes and, hence function.
Materials and Methods
Plant Material
Arabidopsis thaliana (Col-0) was used throughout. The two T-DNA insertion mutants, plc7-3 (SALK_030333) and plc7-4 (SALK_148821) were obtained from the SALK collection1. Homozygous plants were identified by PCR in F2 generation using gene-specific primers. For the identification of plc7-3, we used forward primer 5′-GATTTGGGTGATAAAGAAGTTTGG-3′; reverse primer 5′-CTCCACACAATCTCAGCATTAC-3′ and left border primer LBb1.3 (5′-ATTTTGCCGATTTCGGAAC-3′, in combination with the forward primer). For plc7-4 identification, forward primer 5′-TCCTTCCTGTTATCCATGACG-3′; reverse primer 5′-TTGAAGAAAGCATCAAGGTGG-3′) and left border primer LBb1.3 (in combination with the reverse primer) were used. To generate a plc5/7-double mutant, plc7-3 was crossed with plc5-1 (SALK_144469), a T-DNA insertion KD mutant that was functionally complemented (Zhang et al., 2018b).
Root Growth
Seeds were surface sterilized in a desiccator using 20 ml thin bleach and 1ml 37% HCl for 3 h, and then sowed on square petri dishes containing 30 ml of ½ strength of Murashige and Skoog (½MS) medium (pH 5.8), 0.5% sucrose, and 1.2% Daishin agar (Duchefa Biochemie). Plates were stratified at 4°C in the dark for 2 days, and then transferred to long day conditions (22°C, 16 h of light and 8h of dark) in a vertical position, under an angle of 70°. Four-day-old seedlings of comparable size were then transferred to ½MS-agar plates without sucrose and allowed to grow further for another 6–8 days. Plates were then scanned with an Epson Perfection V700 scanner and primary root length, lateral root number and average lateral root length from each genotype determined through ImageJ software (National Institutes of Health).
Cloning and Plant Transformation
To generate pPLC7::GUS-SYFP reporter line, the PLC7 promoter was amplified from genomic DNA using the following primers: PLC7proH3fw (5′-CCCAAGCTTGATCCTATCAATATTCCTAATTCAGC-3′) and PLC7proNheIrev (5′-CTAGCTAGCTTGAACAATTCCTCAAGTG-3′). The PCR product was cloned into pGEM-T easy and sequenced. A HindIII-pPLC7-NheI fragment was then ligated into pJV-GUS-SYFP, cut with HindIII and NheI. A pPLC7::GUS-SYFP fragment, cut with NotI and transferred to pGreenII-0229. A MultiSite Gateway Three-Fragment Vector Construction Kit2 was used to generate PLC7-overexpression lines, driven by the UBQ10 promotor (pUBQ10::PLC7). Oligonucleotide primers (5′-GGGGACAACGTTTGTACAAAAAAGCAGGCTATGTCGAAGCAAACATACAAAGT-3′ and 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCACAAACTCCAACCGCACAAGAA-3′) including attB1 and attB2 sites, were used to PCR PLC7 from cDNA and was cloned into the donor vector (pDONR207) by using BP Clonase II enzyme mix to create entry clone BOX2. BOX1 was pGEM-pUBQ10 entry clone containing attL4 and attR1 sites. BOX3 was pGEM-TNOS entry clone containing attR2 and attL3 sites. The three entry clones (BOX1, BOX2 and BOX3) and a destination vector (pGreen0125) were used in MultiSite Gateway LR recombination reaction to create the expression clone (Invitrogen).
All constructs were transformed into Agrobacterium tumefaciens, strain GV3101, which was subsequently used to transform Arabidopsis plants by floral dip (Clough and Bent, 1998). Homozygous lines were selected in T3 generation and used for experiments.
RNA Extraction and Q-PCR
The primer pairs to check for PLC1 to PLC9 expression were obtained from Tasma et al. (2008). Similarly, CUC2- and MIR164A-expression levels were determined with the primers described by Bilsborough et al. (2011). The primer pair to measure PLC7 (At3g55940) expression levels was: 5′-GGCTTTCAATATGCAGGGACT-3′ and 5′-CGGGTCAAATAACAGCGTTGG-3′. Total RNA was extracted with Trizol reagent (Invitrogen, Carlsbad, CA). Total RNA (1.5 μg) from 10-day-old seedlings, or 4-week old rosette leaves, were converted to cDNA using oligo-dT18 primers, dNTPs and SuperScript III Reverse Transcriptase (Invitrogen), according to the manufacturer’s instructions. Q-PCR was performed with ABI 7500 Real-Time PCR System (Applied Biosystem). The relative gene expression was determined by comparative threshold cycle value. Transcript levels were normalized by the levels of SAND (At2g28390; forward primer: 5′-AACTCTATGCAGCATTTGATCCACT-3′, reverse primer: 5′-TGAAGGGACAAAGGTTGTGTATGTT-3′; Hong S.M. et al., 2010) or OTC (At1g75330; forward primer: 5′-TGAAGGGACAAAGGTTGTGTATGTT-3′, reverse primer: 5′-CGCAGACAAAGTGGAATGGA-3′) (Bilsborough et al., 2011; Han et al., 2013). Three biological- and two technical replicates were performed for means and standard deviations (Han et al., 2013).
Histochemical GUS Analysis
GUS staining was performed as described previously (Zhang et al., 2018a,b). Briefly, transgenic plants carrying pPLC7::GUS-SYFP were grown for indicated times, after which specific tissues were taken and incubated in an X-Gluc reaction solution containing 1 mg/ml 5-bromo-4-chloro-3 indolyl-β-D-glucuronic acid (X-gluc), 50 mM phosphate buffer (pH 7.0), and 0.1% TX-100. Material was incubated overnight at 37°C and the next day cleared by 70 % ethanol and kept in that solution. GUS staining was visualized under a stereo microscope (Leica MZFLIII) and digitalized with a ThorLabs, CCD camera.
Seed Staining and Sugar Analysis
To visualize seed coat mucilage, mature dry seeds were stained as described in Macquet et al. (2007). Seeds were directly incubated in 0.03% (w/v) Ruthenium red, or after imbibition in 0.5 M EDTA, pH 8.0, for 90 min. For the latter, seeds were washed with water to remove the EDTA and then stained for 20 min with Ruthenium red. Stained seeds were routinely observed with a light microscope (Aristoplan; Leitz). To visualize the surface and the adherent mucilage (AM) layer by confocal microscopy, Calcofluor white (0.01%) and Pontamine S4B were used as staining solutions (Western et al., 2000; Saez-Aguayo et al., 2014). Optical sections were obtained with an Olympus LX81 spectral confocal laser-scanning microscope. A 405 nm diode laser was used to excite Calcofluor white and the emission detected between 412 and 490 nm. For Pontamine S4B, a 561 nm diode laser was used and the detection performed between 570 and 650 nm. For comparisons of the signal intensity within one experiment, the laser gain values were fixed. Three different batches of seeds were analyzed and all of them showed the same phenotype. LUT green fire blue filter and LUT fire filter (Image J) were applied to the Calcofluor white and Pontamine S4B images, respectively.
Soluble carbohydrates were determined as described by Ribeiro et al. (2014), with minor modifications. Three milligrams of dry seeds were ground to powder afterwhich 1 mL of methanol (80% v/v) was added together with 40 μg mL-1 melezitose as internal standard. Samples were incubated in a water bath for 15 min at 76°C before being completely dried by vacuum centrifugation. After addition of 500 μL milliQ water, samples were thoroughly vortexed and centrifuged for 5 min at 17000 ×g. The supernatant was injected onto a Dionex HPLC system (Dionex, Sunnyvale, CA, United States) consisting of a gradient pump module (model GP40), a CarboPac PA100, 4 mm × 50 mm guard column, CarboPac PA100 4 mm × 250 mm seperating column, and an ED40-pulsed electrochemical detector. Soluble carbohydrates were separated by elution with increasing concentrations of NaOH (50–200 mM) at a flow rate of 1 mL min-1. Peaks were identified by co-elution of standards. Quantities were corrected via the internal standard and transformed into μg sugar per mg dry weight.
Leaf-Shape Analysis
Rosettes from 4-week old plants, grown under long-day condition (22°C; 16 h light/8 h darkness), were detached and photographed immediately. Leaves were subsequently removed from the rosette, adhered to white paper using clear adhesive tape and then scanned (Epson Perfection V700 scanner). The 8th leaf was used for calculation. Blade length, -width, -perimeter, -area, -serration number and serration levels were calculated from silhouettes using ImageJ software. Leaf-serration levels are expressed as the distance from tip-to-midvein divided by the distance from sinus-to-midvein, for indicated tooth (2nd–4th) (Kawamura et al., 2010).
Stomatal Aperture
Stomatal aperture measurements were performed according to Distéfano et al. (2012) with minor modifications. Treatments were performed on epidermal strips excised from the abaxial side of fully expanded Arabidopsis leaves of 3-week-old plants, grown at 22°C under 16 h of light and 8 h of dark. Strips were immediately floated onto opening buffer (5 mM MES-KOH, pH 6.1, 50 mM KCl) for 3 h, and subsequently transferred to opening buffer ± ABA. After 90 min, stomata were digitized using a Nikon DS-Fi 1 camera, coupled to a Nikon Eclipse Ti microscope. Stomatal-aperture width was measured using ImageJ software (National Institute of Health).
32Pi-Phospholipid Labeling, Extraction and Analysis
Developing seeds at 10 DAP were carefully removed from the silique. Mature seeds were sterilized and stratified on ½MS (pH 5.8) plates as described, and germinated under long-day conditions for around 20 h when testa ruptured. Both developing and germinating seeds were then transferred to 200 μl labeling buffer (2.5 mM MES, pH 5.8, 1 mM KCl) containing 5–10 μCi 32PO43- (32Pi) (carrier free; Perklin-Elmer) in a 2 ml Eppendorf Safelock tube for 24 h.
Five-day-old seedlings were incubated O/N in labeling buffer and the next day labeled for 3 h. Samples were treated by adding 200 μl labeling buffer ± sorbitol (final concentration, 300 mM) for 30 mins. Labeling and treatments were stopped by adding perchloric acid (final concentration, 5% by vol.) for 5–10 min, after which lipids were extracted and phospholipids separated by TLC (Munnik and Zarza, 2013). Radioactive phospholipids were visualized by autoradiography and quantified by phosphoimaging (Molecular Dynamics, Sunnyvale, CA, United States). Individual phospholipid levels are expressed as the percentage of total 32P-lipids.
Drought Tolerance Assays
Drought assays were performed as described (Zhang et al., 2018a,b). In brief, seeds were stratified at 4°C in the dark and sowed in pots (4.5 cm × 4.5 cm × 7.5 cm) containing equal amounts (80 g) of soil. Nine plants per pot were grown under short-day conditions (22°C with 12 h light/12 h dark) for 4 weeks and then subjected to dehydration by withholding them for water for 2 weeks while control plants were normally watered. Each experiment used 36 plants per genotype and experiments were repeated at least twice. To assay water-loss, rosettes from 4-week-old plants were detached and the FW determined every hour by weighing. Water content was calculated as the percentage from the initial FW. Twenty plants were used for each experiment and independently repeated twice.
Results
Expression of PLC7 During Plant Development
Histochemical GUS analyses on pPLC7-GUS-SYFP reporter lines indicated that PLC7 was mainly expressed in the vasculature throughout all stages of development, including root, cotyledons, leaves, hypocotyl, flower (stamen, style, petal, sepal, receptacle and pedicel) and silique septum (Figure 1), which is similar to the pattern of PLC3 (Zhang et al., 2018a) and PLC5 (Zhang et al., 2018b). After germination, PLC7 expression was mainly observed in the hypocotyl (Figures 1A,B), which then spread to the vasculature throughout the plant upon further development (Figures 1C–I). Interestingly, expression was quite abundant at hydathodes, both in young seedling and mature plants (Figures 1B–E,L, indicated by arrows). Unlike PLC3 and PLC5, PLC7 did not display the characteristic, “segmented” expression in the root vasculature. Instead, expression was homogenous in both main root- and lateral root vasculature (Figures 1F,G), and stopped near the transition zone (Figure 1H). GUS staining was also visible in trichomes (Figures 1J,K), similar to PLC5 but stronger, and different from PLC3, which only showed expression at the base of the trichome (Zhang et al., 2018a,b). In contrast to PLC3 and PLC5, no GUS activity was detected in guard cells (Figure 1O). We also tried to image the YFP signal but this was unfortunately too low.
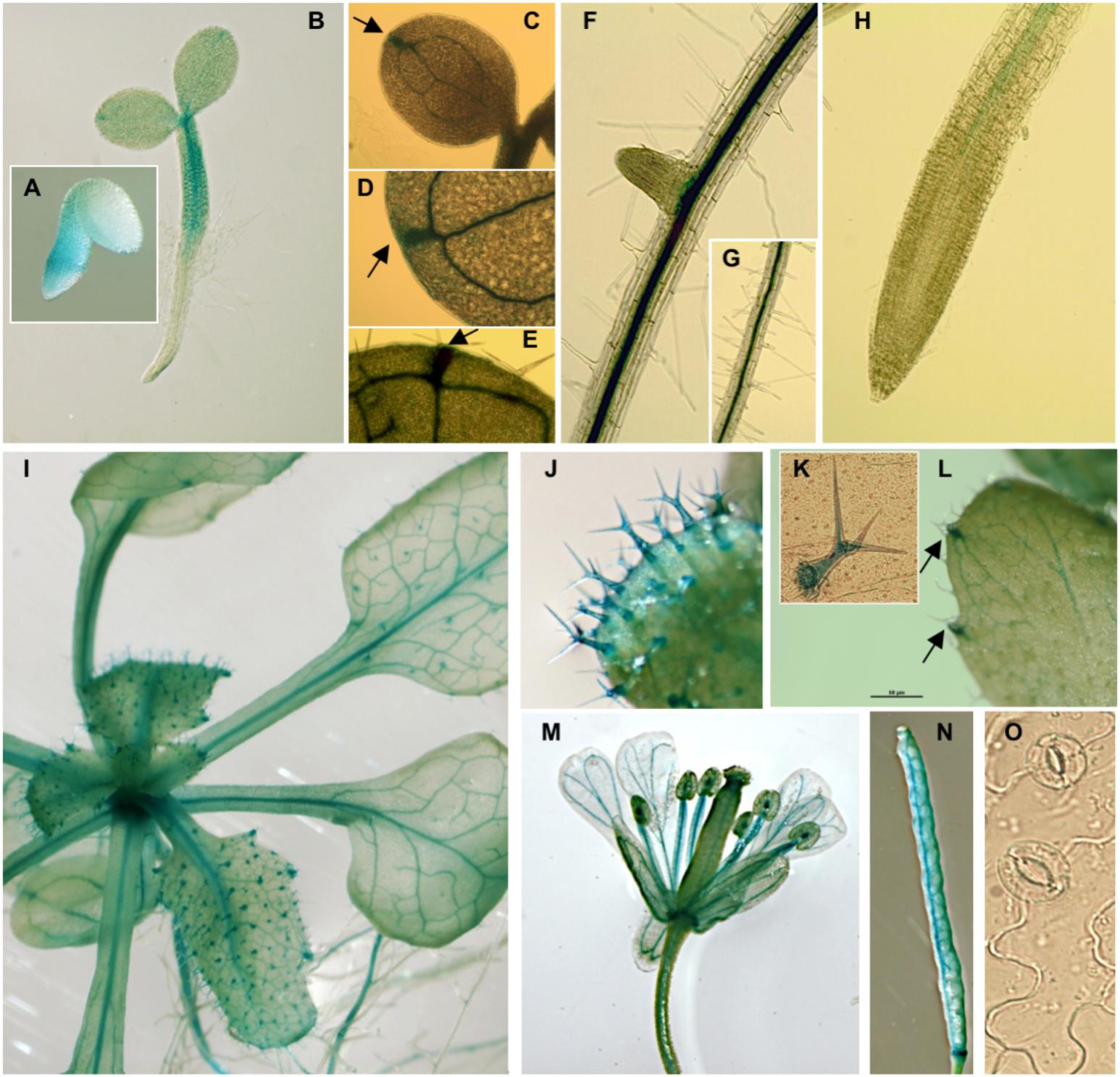
FIGURE 1. pPLC7-GUS expression in seedlings and specific tissues of Arabidopsis. (A) Embryo, 28 hrs after stratification, (B) vascular tissue of 2-d old seedlings, (C–H) 10-d old seedlings, including cotyledons and roots, (I) 3-week-old mature plant, (J,K) trichomes, (C–E,I,L) hydathodes (indicated by arrows), (M) flower, including style, filament, receptacle and pedicel, (N) silique, (O) guard cells (no staining detectable).
While our results confirm the Q-PCR data from Tasma et al. (2008), that PLC7 is expressed throughout the plant, our results indicated that this expression is primarily restricted to the vasculature, hydathodes and trichomes.
plc7 and plc5/7 Mutants Show Wild Type Root Growth
To functionally address the role of PLC7, two homozygous T-DNA insertion lines, plc7-3 (SALK_030333) and plc7-4 (SALK_148821) were obtained (Figure 2A). PLC7 expression was validated by Q-PCR and revealed that plc7-3 is a KO- and plc7-4 a KD mutant (Figure 2B). In contrast to plc3- (Zhang et al., 2018a) and plc5 mutants (Zhang et al., 2018b), the root architecture of plc7 mutants did not differ from wild type (Figures 2C,D).
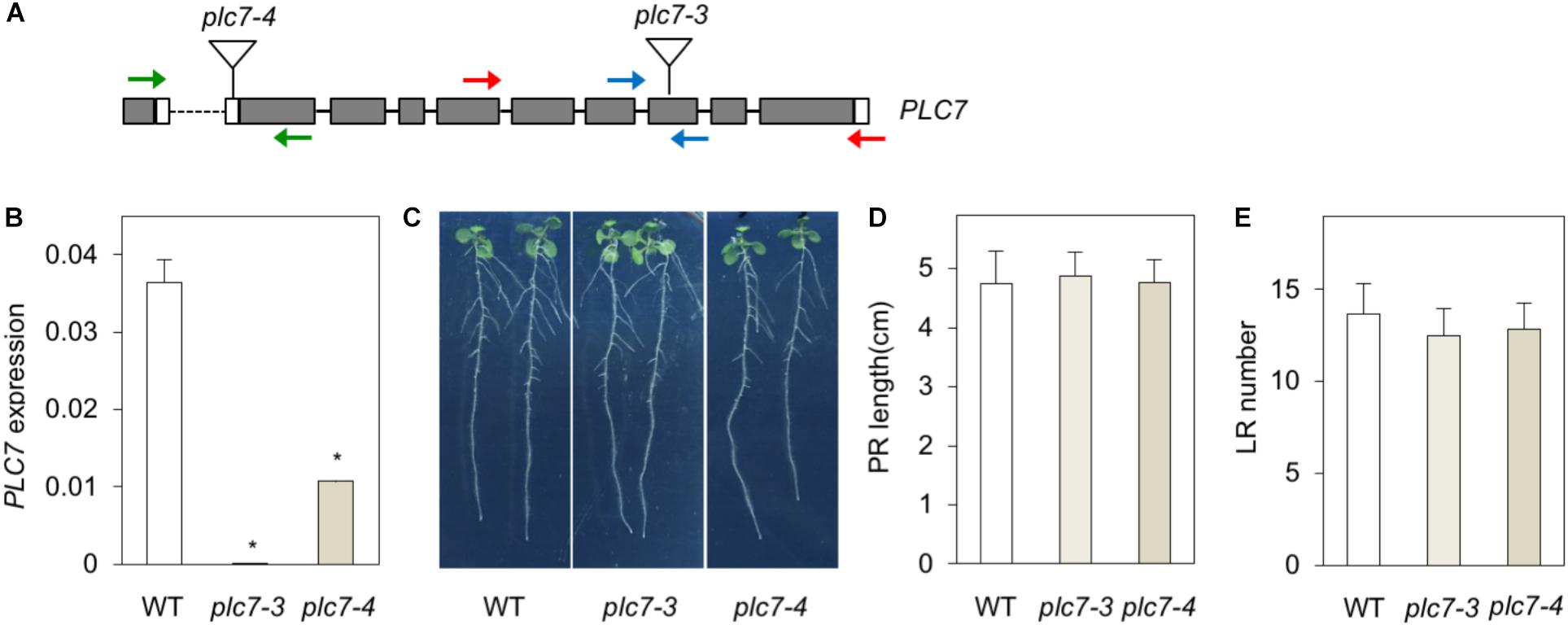
FIGURE 2. KO- or KD of PLC7 does not affect root development. (A) T-DNA insertion positions (triangles) in the PLC7 gene of the plc7-3 (KO) and plc7-4 (KD) lines. Filled boxes and lines represent exons and introns, respectively, while open boxes represent untranslated regions. Blue primers were used for qPCR analyses, red primers for genotyping plc7-3, and green primers for genotyping plc7-4. Forward primer PLC7-4 is located at the last exon and 3’UTR of the gene in front. (B) PLC7 expression levels in wild-type, plc7-3 and plc7-4 measured by Q-PCR using SAND as a reference gene. Values are the means ± SD (n = 3) of a representative experiment that was independently repeated three times. (C) Seedling morphology of wild type and plc7 mutants. Seeds were germinated on ½MS with 0.5% sucrose for 4 days and then transferred to ½MS plates without sucrose. Photographs were taken 12 days after germination (DAG). (D) Primary root (PR) length and (E) lateral root (LR) number at 12 DAG. Values are means ± SE of three independent experiments (n > 20). ∗Indicates significance at P < 0.05 compared to wild-type, based on Student’s t-test.
To analyze gene redundancy, we tried to generate plc3/5/7-triple mutants by crossing plc7-3 with the plc3/5-double mutant (Zhang et al., 2018b). After genotyping T2- and T3 populations, we could not identify any plc3 plc7-double mutants nor any homozygous triple mutants. We did find homozygous plc5/7-double mutants (Supplementary Figure S1) but as shown in Supplementary Figures S1B–D, no significant differences between the root systems of plc5/7- and wild-type seedlings were found.
plc5/7 Mutant Displays Mucilage Defect
While imbibing seeds for stratification, we noticed that the volume of the plc5/7-seed pellet was always smaller than wild-type’s after O/N incubation (Figure 3A). This was not the case for the individual plc7 or plc5 mutants (Supplementary Figure S2; Zhang et al., 2018b). Upon imbibition, the seed coat-epidermal cells normally extrude mucilage that forms two transparent layers (adherent and non-adherent layers) around the seed. To examine whether the smaller volume of the plc5/7 mutant was caused by a mucilage defect, we stained imbibed seeds with Ruthenium red (Figure 3B) that stains pectins, the main component of mucilage (Golz et al., 2018). Compared to wild-type, the adherent and non-adherent layers were more expanded in the plc5/7 mutant (Figure 3B, top panel) and when seeds were mildly shaken to remove the non-adherent layer, or treated with EDTA, plc5/7 seeds lost their adherent layer completely (Figure 3B, middle and lower panel, respectively).
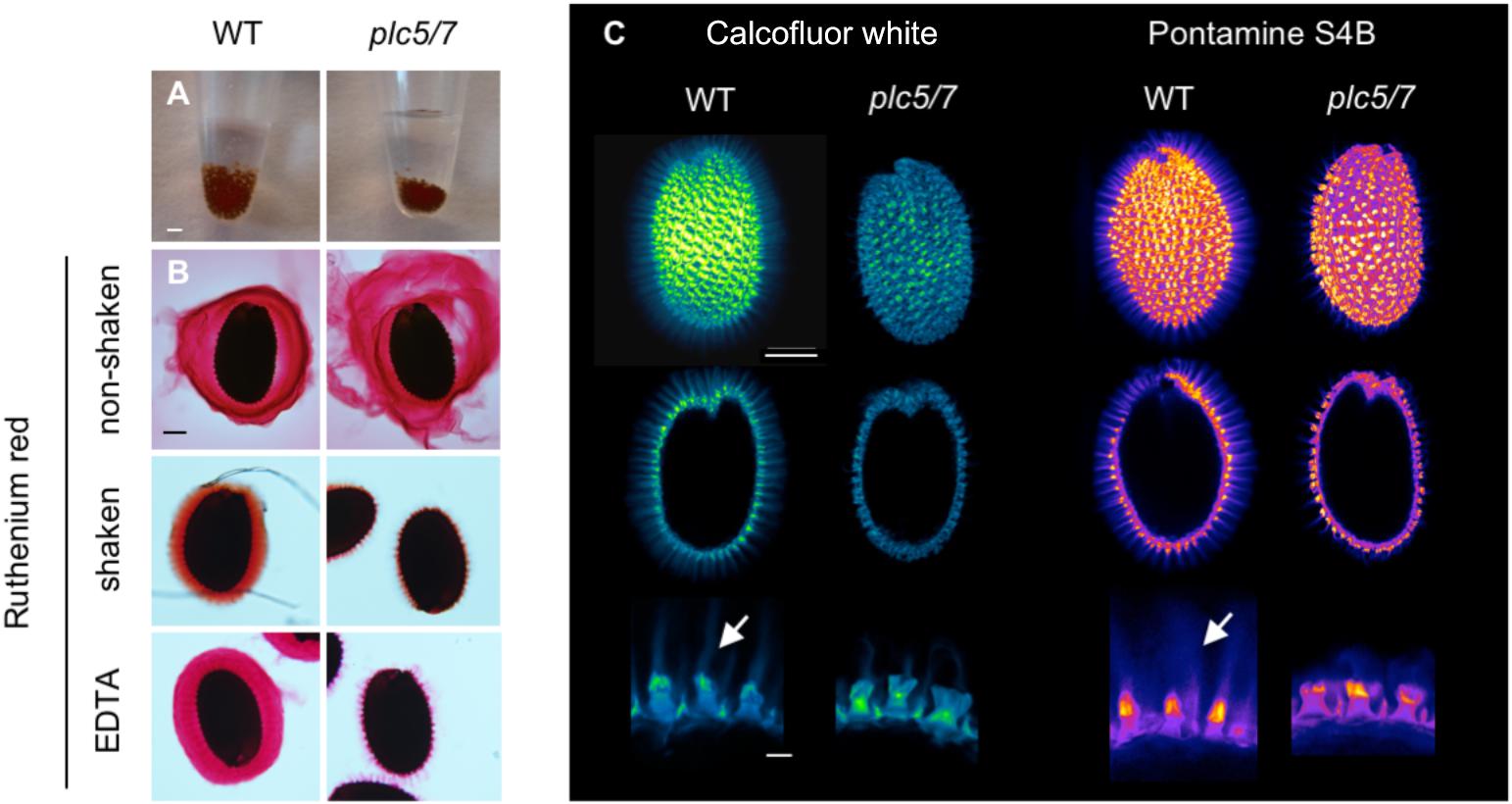
FIGURE 3. Seeds of plc5/7-double mutant exhibit a mucilage defect. (A) Seeds of plc5/7 swell less than WT during imbibition. Equal amounts of dry seeds of WT and mutant were immersed in water O/N and photographed the next day. (B) Ruthenium red staining of wild type- and plc5/7 seeds without shaking (top), with shaking (middle), or EDTA treatment (bottom). ’With’ shaking shows adherent mucilage layer; ’without’ shaking displays both adherent and non-adherent mucilage layers. (C) Cellulose staining by Calcofluor white (left panel) or Pontamine B (right panel) in wild type- and plc5/7 seeds. Confocal images of whole seeds, cross section, and close-up views (top, middle and bottom, respectively) are shown. Bars represent 2 mm (A), or 0.1 mm (top and middle rows) or 0.025 mm (bottom row) (B). Representative results of at least 3 biological replicates are shown.
Increased solubility of the pectins has been linked to perturbation of cellulose deposition (Basu et al., 2016; Ben-Tov et al., 2018). To test this, wild type- and plc5/7 seeds were stained with Calcofluor White (CFW, for cellulose and other β-glucans staining; (Figure 3C, left panel) or Pontamine S4B (cellulose-specific dye; Figure 3C, right panel) (Anderson et al., 2010; Wallace and Anderson, 2012). In wild-type seeds, the primary cell wall remnants and rays extending from the columella were stained by both dyes (Figure 3C). The staining pattern of plc5/7 seeds appeared similar, but the CFW intensity was lower and the rays were clearly reduced compared to wild-type. These results point to a role for PLC5 and PLC7 in cellulose-ray formation, which is a novel function for PLCs.
PLC5 and PLC7 Are Expressed in Developing Seeds
To investigate the expression of PLC5 and PLC7 during seed development, additional histochemical GUS analyzes were performed (Figure 4). At 4 days after pollination (DAP), some GUS activity was found for PLC5 (Figure 4A) while a strong staining in the seed coat and chalazal area for PLC7 was obtained (Figure 4B). Later in development (8 and 10 DAP), expression increased, with PLC5 expression appearing in the seed coat and funiculus (Figure 4A), and PLC7 becoming stronger in the seed coat and chalazal (Figure 4B).
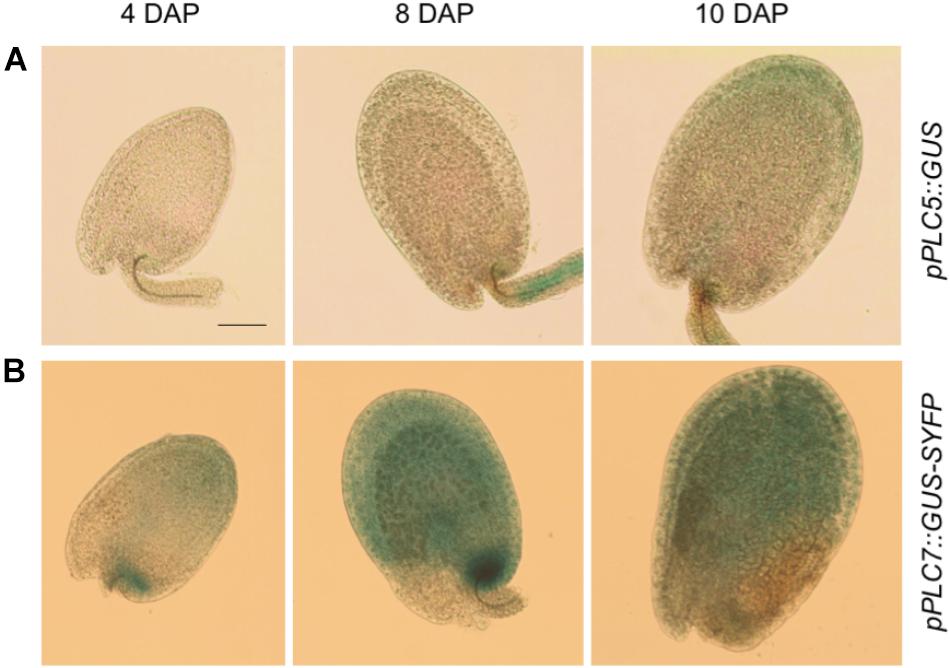
FIGURE 4. PLC5 and PLC7 expression during seed development. (A) GUS activity analysis in pPLC5::GUS-developing seeds. Expression was found in the funiculus at 8 days after pollination (DAP) and in the seed coat at 10 DAP. (B) GUS activity analysis in pPLC7::GUS-SYFP-developing seeds. Staining was found in the chalazal and seed coat. Representative results of 3 independent experiments are shown. Bar = 0.1 mm.
PPI- and PA Levels in Developing and Germinating Seeds
To analyze substrate- (i.e., PIP and PIP2) and product- (conversion of PLC-generated DAG into PA) relationships, wild type- and plc5/7 seeds at 10 DAP were compared with germinating, mature seeds after 24 h of 32Pi-labeling. As shown in Figure 5, wild-type and plc5/7 seeds contained similar amounts of PIP2, PIP and PA in both stages. Interestingly, PIP- and PA levels were much higher in developing seeds, while PIP2 levels were similar in both stages.
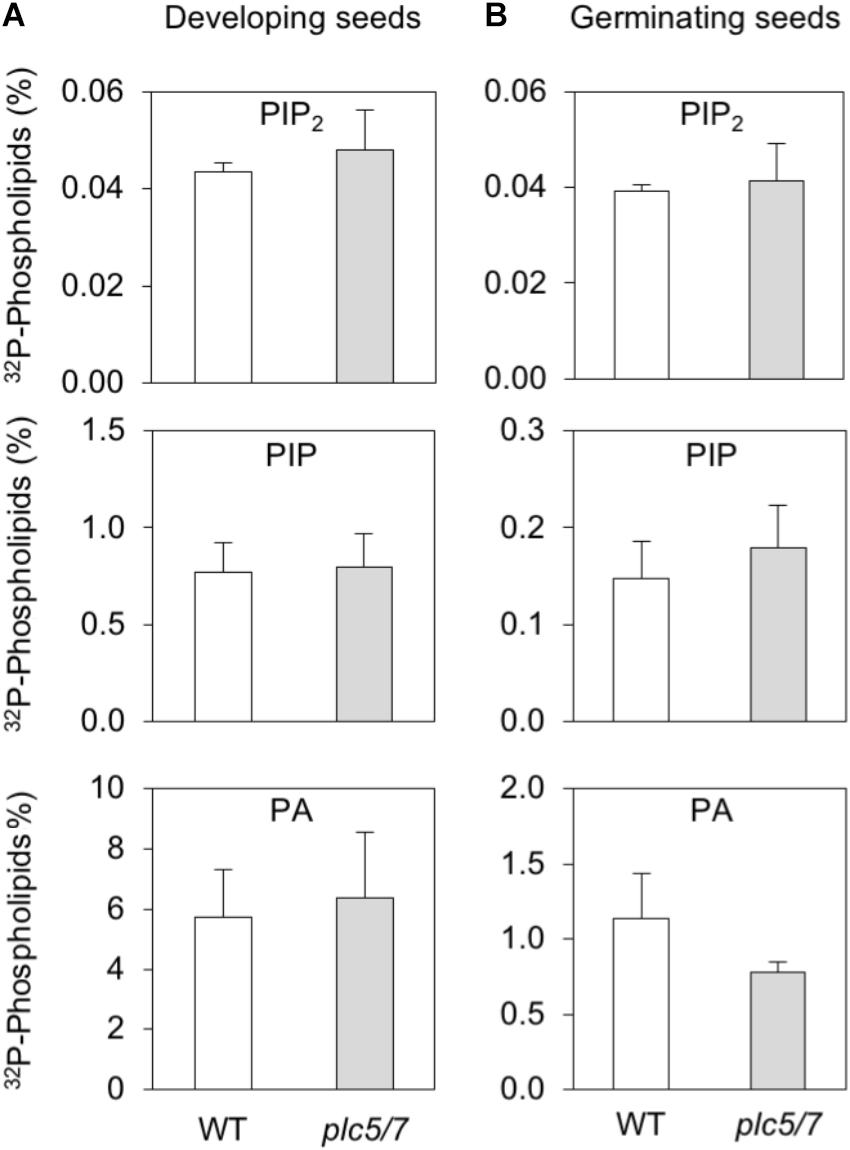
FIGURE 5. PPI- and PA levels in developing- and germinating (mature) seeds of wild type and plc5/7. (A) Developing seeds (∼00; 10 DAP) or (B) pre-germinated mature seeds (∼200) of wild type and plc5/7 were labeled with 32Pi for 24 h and their lipids extracted, separated by TLC and quantified by Phosphoimaging.32P-levels of PIP2, PIP and PA are expressed as percentage of total 32P-phospholipids. Three independent experiments were performed; data shown are means ± SD (n = 3) of a representative experiment.
Enhanced Leaf Serration Phenotype for plc5/7 Plants
Growing plc5/7 mutants on soil revealed a novel phenotype, i.e., the patterning of their leaf-edge (serration). This phenotype was absent from the individual plc7 or plc5 mutants (Supplementary Figure S3; Zhang et al., 2018b). Overall, the level of serration in successive rosette leaves was significantly increased in plc5/7, which appeared to be stronger in the proximal part of the blade than in the distal part (Figures 6A,B). To quantify this in more detail, we measured various parameters of the 8th leaf (Figures 6C–E) of 4-weeks old rosettes of both genotypes. No changes in blade length were observed between wild type and plc5/7. Blade width, -perimeter and -area, appeared to be slightly bigger in plc5/7 but this was not significant (Figure 6D). The serration number neither changed, but the serration level (indicated by the ratio between the distance from the midvein to tip and the distance from the midvein to sinus; Figure 6C) was significantly higher in three successive teeth (Figure 6F).
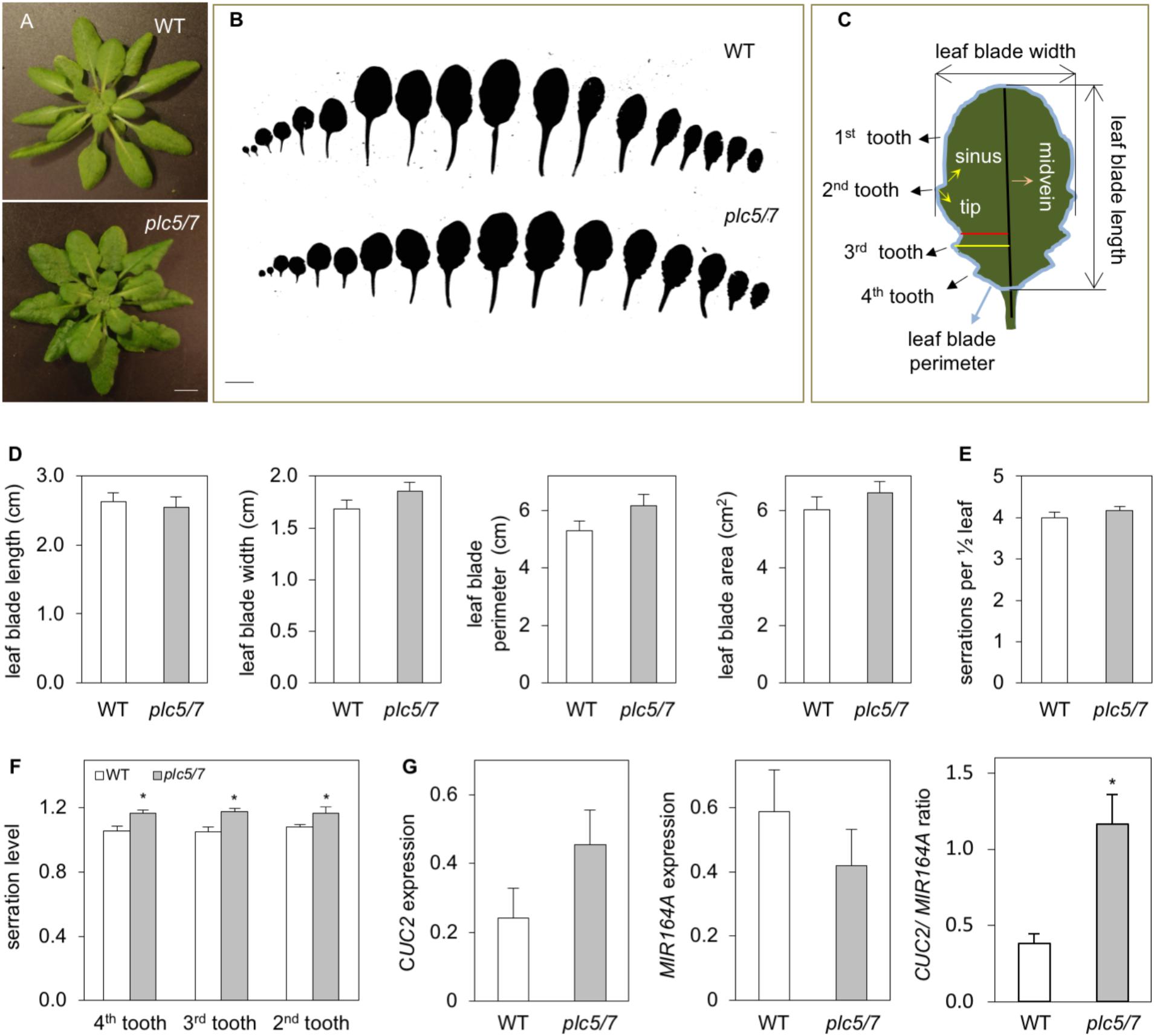
FIGURE 6. Leaves of plc5/7 plants display enhanced leaf serration. (A) Rosette of wild type and plc5/7 mutant. Plants were grown on soil for 4 weeks in short-day conditions, afterwhich rozettes were cut and photographed immediately. Bar = 1 cm. (B) Leaf series of 4-week old wild type and plc5/7 plant. (C) Cartoon demonstrating the leaf parameters measured of the 8th leaf. (D) Quantification of blade size including, length, width, perimeter and area. (E,F) Quantification of leaf serration number (E) and level (F) in wild type and plc5/7 mutant. (G) Expression of CUC2 and MIR164A and their ratio in WT and plc5/7 mutant, relative to the expression of reference gene, OTC. Data represents the means ± SD (n = 3) from a representative experiment that was repeated twice with similar results. Asterisk (∗) indicate significance at P < 0.05 compared to WT, based on Student’s t-test.
In Arabidopsis, leaf-margin development is controlled by a balance between microRNA164A (MIR164A) and CUP-SHAPED COTYLEDON2 (CUC2) (Nikovics et al., 2006). Hence, we compared the expression of MIR164A and CUC2 in wild type and plc5/7 leaves. As shown in Figure 6G, plc5/7 leaves were consistently found (three independent experiments) to contain higher levels of CUC2 and lower levels of MIR164A, resulting in a significant increase in the CUC2/MIR164A ratio.
plc5/7 Mutants Are Better Protected Against Drought
When plants were left in the greenhouse without watering, we noticed that plc5/7 mutants appeared to be more drought tolerant while single mutants behaved like wild type (data not shown). Performing multiple drought assays confirmed this (Figure 7A), while detached rosettes of 4-week-old plc5/7 plants lost less water than wild type (Figure 7B).
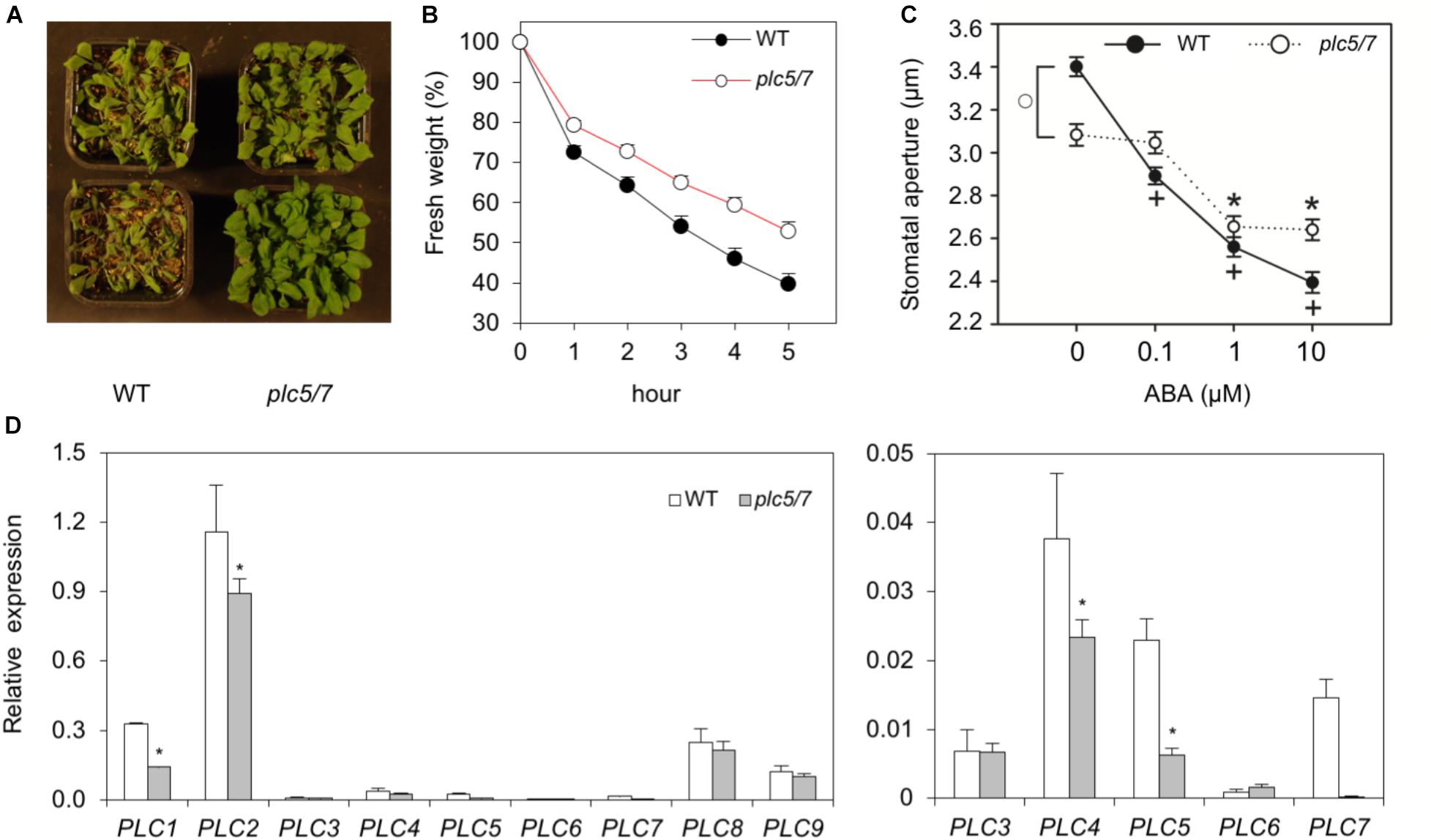
FIGURE 7. Soil grown-plc5/7 plants are more tolerant to drought stress. (A) Six-weeks old plants of wild type- or plc5/7, grown on soil and exposed to drought by withholding water for the last 2 weeks. (B) Water loss of detached rosettes of normally watered, 4-weeks old plants. Water loss was measured at indicated time points and expressed as a percentage of the initial fresh weight. Values are means ± SD for one representative experiment for 3 independent experiments (n = 36). (C) Effect of ABA on the stomatal aperture in leaf strips of wild type and plc5/7 plants. Values are means ± SE of at least three independent experiments (n > 100). (D) Left panel: PLC1- PLC9 expression levels in wild type- and plc5/7 leaves measured by Q-PCR, relative to SAND expression. Right panel: Zoom-in of the expression of PLC3-PLC7. Values are means ± SD (n = 3) for a representative experiment that was repeated twice with similar results.
ABA plays a key role during the response to dehydration stress and is known to induce stomatal closure to reduce water loss (Sean et al., 2010). Hence, we checked the stomatal-closure of plc5/7 and wild type in response to ABA. As shown in Figure 7C, plc5/7 has less-open stomata compared to wild type without ABA, while upon ABA treatment, the plc5/7 mutants were less responsive.
Previously, we showed that overexpression of PLC3 or PLC5 enhanced drought tolerance in Arabidopsis (Zhang et al., 2018a,b), which was found earlier for maize, canola and tobacco (Wang et al., 2008; Georges et al., 2009; Tripathy et al., 2011). We therefore wondered whether the increase in drought tolerance in plc5/7 was a result of the overexpression of any of the other (redundant) PLCs. However, no strong overexpression of any PLC was found in plc5/7 (Figure 7D). In fact, PLC1, PLC2 and PLC4 appeared to be slightly down-regulated (Figure 7D).
Overexpression of PLC7 Increases Drought Tolerance
As mentioned above, overexpression of Arabidopsis PLC3 or PLC5, which are from different subfamilies than PLC7, resulted in enhanced drought tolerance (Zhang et al., 2018a,b). To check the effect of the overexpression of PLC7, homozygous T3 plants were generated. Two lines, PLC7-OE9 and PLC7-OE12, which overexpressed PLC7 around 80- to 100-fold, respectively, were selected for further studies (Figure 8A). Both lines were found to be more drought tolerant than WT (Figure 8B), and lost slightly less water when rosettes of 4-week-old plants were detached (Figure 8C). The stomatal aperture and their response to ABA was found to be similar to wild type (Figure 8D), which is different from PLC3- and PLC5-OE plants that had more closed stomata than wild type.
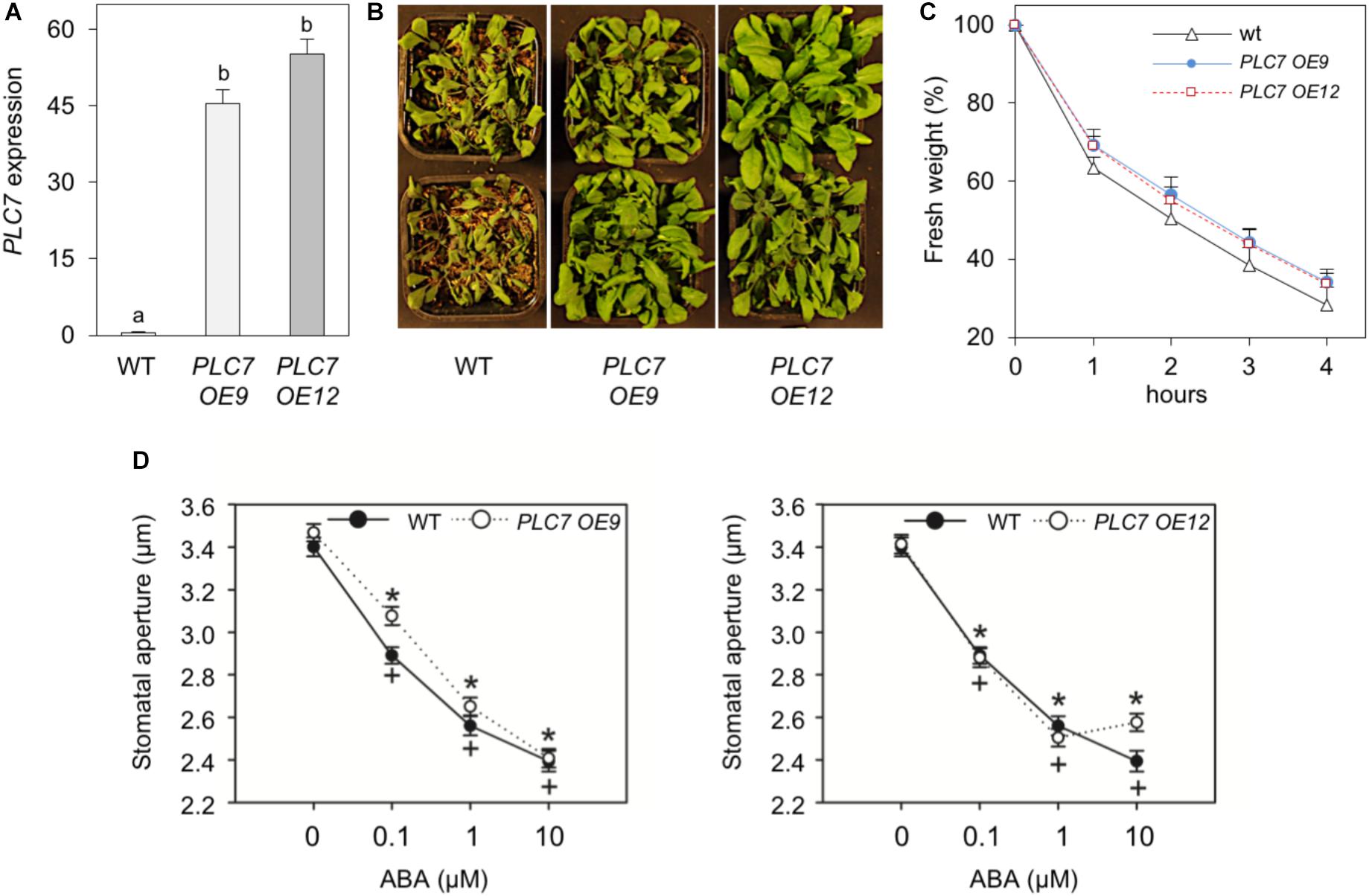
FIGURE 8. Overexpression of PLC7 enhances drought tolerance. (A) PLC7 expression levels in wild type and two homozygous PLC7-overexpression lines, PLC7-OE9 and PLC7-OE12 as measured by Q-PCR relative to SAND. Values are means ± SD (n = 3) for one representative experiment. (B) Phenotype of 6-week old plants from wild type and PLC7-overexpresion lines, #OE9 and #OE12, after 2 weeks of drought. (C) Water loss of detached rosettes of 4-weeks old plants. Values are means ± SD for one representative experiment for 3 independent experiments (n = 36). (D) Stomatal aperture in leaf peels of wild type, PLC7-OE9 (left), PLC7-OE12 (right) and the effect of ABA. Values are means ± SE of at least three independents (n > 100).
Phospholipid Responses in Osmotically Stressed Seedlings
To analyze phospholipid responses in plc5/7 and the PLC7-OE lines, 32P-labeling experiments (3 h pre-labeling) were performed on seedlings and the effect of sorbitol tested to mimic osmotic stress. Both plc5/7 and wild type showed similar PPI- and PA levels in the absence of sorbitol (Figures 9A,B). Upon sorbitol treatment, a consistent stronger PIP2 response was observed for plc5/7 seedlings in all three independent experiments (P-value almost 0.05). While PIP2 levels increased by ∼4 times in wild type, in plc5/7 seedlings a typical 6-times increase was found. No such differences in PA- or PIP were observed (Figure 9B).
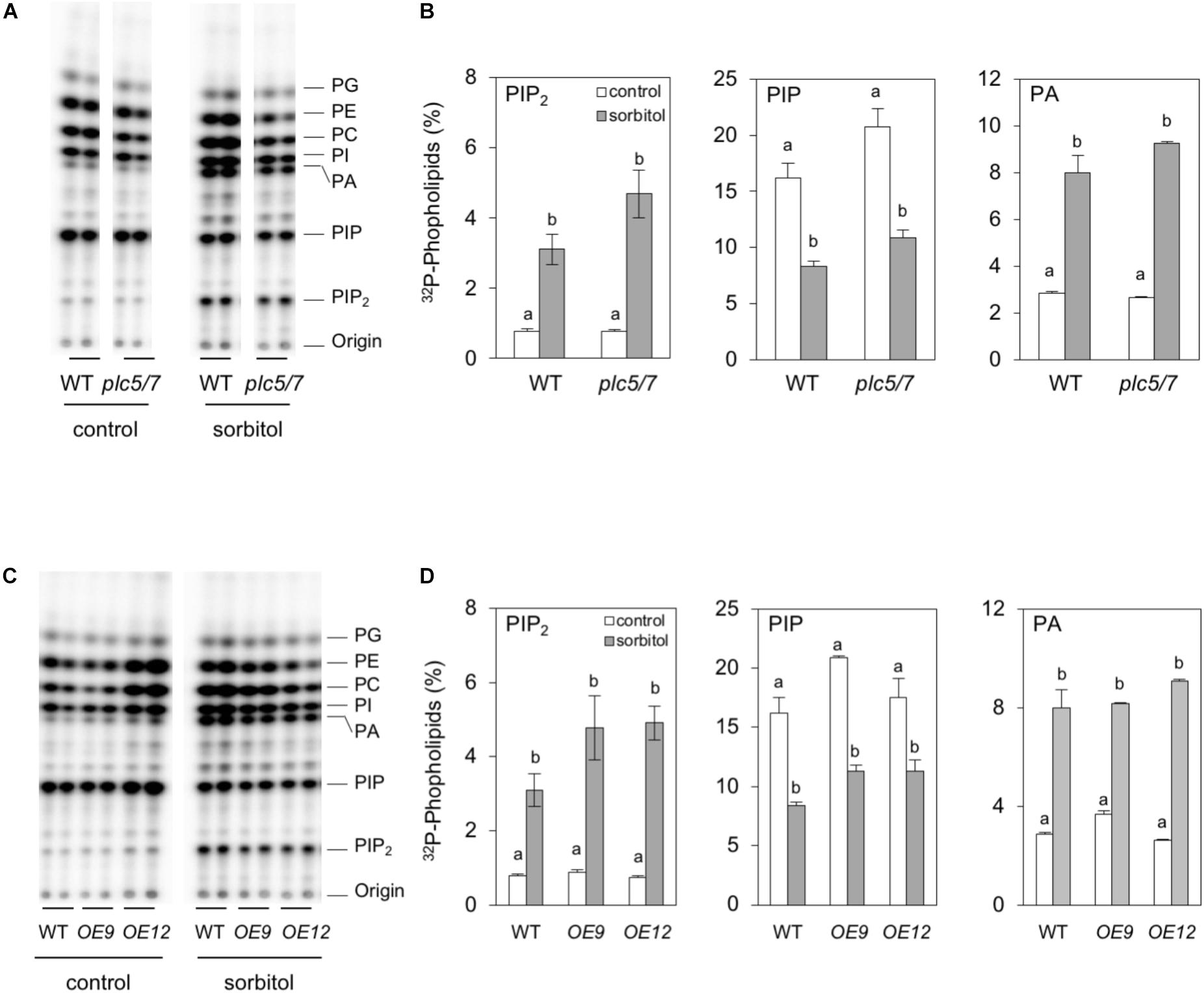
FIGURE 9. Hyperosmotic stress triggers stronger PIP2 responses in plc5/7- and PLC7-OE seedlings than WT. Six-day-old seedlings were 32P-labeled for 3 h and then treated with or without 300 mM sorbitol for 30 min. Extracted lipids were analyzed by TLC and quantified through phosphoimaging. (A,B) WT vs plc5/7 seedlings, (C,D) WT vs PLC7-OE lines #9 and #12. (A,C) Typical TLC profiles, (B,D) 32P-levels in PIP2, PIP and PA. Data shown are the means ± SE (n = 3) of one experiment, representative of three independent experiments. Data was analyzed by 2-way ANOVA. Statistical significant differences between normal and sorbitol conditions in wild type and plc5/7 or PLC7-OEs are indicated by letters (P < 0.05).
PLC7-OE lines revealed no difference in PPI- or PA levels compared to wild-type under control conditions (Figures 9C,D). However with sorbitol, again a stronger PIP2 response was observed in PLC7-OE lines, i.e., ∼6-times vs. ∼4-times increase for WT. PA- and PIP responses were similar to wild type (Figures 9C,D). These results suggest that both plc5/7 and PLC7-OE plants boost more PIP2 in response to osmotic stress than wild-type, similar to what we found earlier for PLC3- and PLC5-OE lines (Zhang et al., 2018a,b).
Discussion
Knockout of PLC7 Does Not Affect Root Architecture
Previously, we demonstrated that Arabidopsis PLC3 and PLC5 were both involved in lateral root formation, but that the phenotype in a plc3/5-double mutant was not worse than the individual, single mutants. Hence, we speculated that another PLC might be involved (Zhang et al., 2018a,b). Since promotor-GUS analyses of PLC3 and PLC5 revealed specific expression in phloem companion cells and showed a typical ‘segmented’ pattern in the vascular of the primary root from which lateral roots emerged, we searched for other phloem-specific PLCs. Using the eFP browser, we found two potential candidates, PLC2 and PLC7. Since T-DNA insertion mutants of PLC2 were lethal (Li et al., 2015; D’Ambrosio et al., 2017; Di Fino et al., 2017), we focused on PLC7. Two independent homozygous T-DNA insertion mutants were obtained, with plc7-3 being a KO- and plc7-4 a KD line. Both mutants, however, exhibited normal root architecture (Figure 2). In an attempt to create double- and triple mutants of the potentially redundant PLCs, we crossed plc3-2/plc5-1 (plc3/5; Zhang et al., 2018b) with plc7-3, however, this only resulted in viable plc5/7 double mutants, as the combination plc3/plc7 turned out to be lethal (not shown). The plc5/7 double mutant, however, did not reveal significant changes in root morphology (Figure 2 and Supplementary Figure S1). While pPLC7-GUS analyses confirmed the vascular expression of PLC7, which according to the Arabidopsis eFP Browser (Winter et al., 2007) is all phloem and phloem companion cells, it lacked the typical segmented pattern as found for PLC3 and PLC5 (Figure 1; Zhang et al., 2018a,b). These results may indicate that PLC2 and PLC3 represent redundant PLCs in root development. Interestingly, PLC7 was also expressed in hydathodes and in seeds, which correlates well with the two new phenotypes that were found for plc5/7-double mutants, and have never been observed before. These include a mucilage phenotype in seeds and a serration phenotype in leaves. The latter may correlate with PLC7’s specific expression at the hydathodes. PLC7 was also strongly expressed in trichomes. PLC5 is also expressed in trichomes, although less, and PLC3 is typically expressed at the basal cells of trichomes in developing leaves (Zhang et al., 2018a,b). We checked for trichome phenotypes in individual plc3, plc5 or plc7 and plc3/5- and plc5/7- mutants, but found no obvious differences (number, shape). Again, this could be due to redundancy. Without defects, the role of PLC in trichomes remains unclear.
Another new finding for PLC loss-of-function mutants is that plc5/7 mutants were more drought tolerant, a phenotype that is typically found when PLC is overexpressed (Wang et al., 2008; Georges et al., 2009; Tripathy et al., 2011; Zhang et al., 2018a,b). No upregulation of redundant PLCs in the plc5/7 background was found so it must be a consequence of the plc5/7 combination, possible in combination with the slight down regulation of PLC1, PLC2 and PLC4 that was found. RNASeq analyses of the plc mutant- and OE lines may shed light on potential pathways that are up- or down-regulated to explain various phenotypes. Most importantly, altering expression of PLC genes results in clear defects, which will help elucidating the roles PLC can play in plant signaling and development.
While new phenotypes provide new pieces of the PLC-signaling puzzle, it remains unclear how this is achieved at the cellular and molecular level. Flowering plants lack the prime targets for IP3 and DAG, but there are indications that plant responses are coupled via inositol polyphosphates (IPPs) and/or PA (Munnik, 2001; Arisz et al., 2009, 2013; Munnik and Zarza, 2013; Testerink and Munnik, 2011; Gillaspy, 2013; Munnik, 2014; Laha et al., 2015; Heilmann, 2016a,b; Hou et al., 2016; Noack and Jaillais, 2017; Yao and Xue, 2018). Alternatively, since PIP or PIP2 are emerging as second messengers themselves, PLC could function as an attenuator of signaling (Gillaspy, 2013; Munnik, 2014). In that respect, it is interesting to notice that the increased drought-tolerant phenotype in plc5/7 and PLC3, -5, and -7 overexpression lines correlates well with stronger PIP2 responses upon osmotic stress. How this works remains unclear, however. Maybe they are more primed to enhanced PIP2 turnover.
Role for PLC5 and PLC7 in Seed Mucilage
The mucilage extrudes from seed coat epidermal cells when seeds are exposed to water, which helps seeds to remain hydrated while the germination process is in progress (Western et al., 2000). The major component of mucilage is pectin, of which polygalacturonic acid (PGA) and rhamnogalacturonan I (RGI) are the most common compounds (Carpita and Gibeaut, 1993; Cosgrove, 1997). In addition, several other polysaccharides can be found (containing arabinose, galactose, glucose, xylose and mannose), and are equally important in determining mucilage’s properties (Voiniciuc et al., 2015a,b). Two layers of mucilage can be distinguished, a water- soluble non-adherent outer layer and an adherent inner layer (Zhao et al., 2017). While the non-adherent outer layer is easily removed from the seed, the latter is relatively hard to detach, even chemically (Zhao et al., 2017). The plc5 plc7 double mutant releases mucilage that is less adherent to seeds than in the wild type (Figure 3). In addition, cellulosic rays stained with calcofluor white and pontamine S4B appear shorter around the mutant seeds. These defects have been reported in mutants that disrupt cellulose synthesis (Mendu et al., 2011; Sullivan et al., 2011; Griffiths et al., 2015; Griffiths and North, 2017; Ben-Tov et al., 2018), as well as in mutants that directly impair the synthesis of xylan (Voiniciuc et al., 2015a,b; Hu, 2016; Hu et al., 2016; Ralet et al., 2016). Therefore, PLC could influence membrane phospholipids that are important for the trafficking of cellulose synthase enzymes or the secretion of matrix polysaccharides (such as xylan) from the Golgi apparatus. We checked for sugar-composition aberrations in whole seeds, but could not find changes compared to wild type (Supplementary Figure S4). Possibly, the distribution between non-adherent and adherent mucilage layers might be different, which needs to be further determined.
Histochemical analysis of the GUS-reporter lines indicated that both PLC5 and PLC7 are expressed during seed development (Figure 4). Until now, no mucilage deficiency has been linked to PLC, and we only observed the mucilage defect in the plc5/7-double mutant of all our PLC knockout mutants. The defect in both PLC5 and PLC7 probably breaks the balance for mucilage maintenance by altering cellulose deposition and/or crystallization in the inner mucilage. How the enzyme PLC could be involved in all this will require additional research. Potentially, PLC could be required for mucilage secretion or for the localization or activity of cellulose synthases, for example. PA, PIP and PIP2 have been implicated to play essential roles in vesicular trafficking, -fusion and -fission. Even though no difference was found in PPI- or PA levels in either developing or mature seeds (Figure 5), reduced amounts of PLC5 and lack of PLC7 might cause crucial local changes in lipid or IPP molecules.
Role for PLC in Leaf Serration
Leaf shape is defined by the pattern- and degree of indentation at the margin area, distinguishing many plant species (Tsukaya, 2006). The patterning involves a complex cross-talk between hormone signaling and genetic regulators (Byrne, 2005; Fleming, 2005). The development of leaf serration involves auxin maxima at the protrusion of each serrated section (Hay and Tsiantis, 2006). Genetic studies identified the auxin efflux carrier PIN-FORMED 1 (PIN1) and CUP-SHAPED COTYLEDON 2 (CUC2) as two key factors required (Hay et al., 2006; Nikovics et al., 2006). PIN1 asymmetrically localizes on plasma membranes and directionally transports auxin, creating auxin maxima that direct the outgrowth of the serrations (Hay et al., 2006; Scarpella et al., 2006). CUC2 is a transcription factor that is post-transcriptionally downregulated in leaves by MIR164A (Nikovics et al., 2006). CUC2 expression is limited to the sinus where the serration starts, and the promotion of serration outgrowth is through cell division, not by suppression of sinus growth (Kawamura et al., 2010). CUC2 is also thought to regulate the polarized localization of PIN1 in convergence points at the leaf margin, where it may play a role in establishing, maintaining and/or enhancing auxin maxima that result in leaf serration (Bilsborough et al., 2011; Kawamura et al., 2010). In a feedback loop, auxin downregulates CUC2, both transcriptionally and post-transcriptionally through activation of MIR164A (Bilsborough et al., 2011).
The plc5/7-double mutant revealed a mildly-enhanced leaf-serration phenotype (Figure 6). Subsequent measurement of CUC2- and MIR164A expression revealed an up-regulation of CUC2 and down-regulation MIR164A, consistent with enhanced serration ( Kawamura et al., 2010; Bilsborough et al., 2011). Promotor-GUS analyses showed that both PLC5 and PLC7 were expressed at leaf hydathodes, a secretory tissue that secretes water through the leaf margin that is associated with leaf serration (Tsukaya and Uchimiya, 1997) and auxin response maxima (Scarpella et al., 2006). Hence, it is possible that PLC5 and PLC7 redundantly contribute to the regulation of leaf serration, since the phenotype was absent in the single mutants. We also measured PPI-and PA levels in plc5/7 rosette leaves, but like seeds and seedlings, we found no significant changes (data not shown; Figures 5, 9). It is possible that only small changes occur in particular cells and tissues but that most have normal levels/responses so that differences are lost in the total background. How PLC5 and PLC7 are involved in leaf serration requires further investigation.
Role for PLC in Drought Tolerance
Earlier, overexpression of PLC in maize, tobacco and canola have been shown to improve their drought tolerance (Wang et al., 2008; Georges et al., 2009; Tripathy et al., 2011). Similarly, overexpression of PLC3 or PLC5 in Arabidopsis was shown to improve their drought tolerance (Zhang et al., 2018a,b), and here we show this also holds for PLC7. Interestingly, we also found an increased drought-tolerant phenotype for plc5/7-double mutants, which did not occur in the single plc5-1 and plc7-3 mutants. Previous results showed that PLC3-OE and PLC5-OE lines showed a reduced closing response to ABA compared to wild type and had less stomata open in the absence of ABA (Zhang et al., 2018a,b). Under control conditions, overexpression of PLC7 did not reveal this “less-open stomata” phenotype, and their stomatal response to ABA was similar to wild type. Stomata of plc5/7 plants were less open, which is in contrast to the plc5- and plc7-single mutants that had a normal opening at control conditions (Figure 7C and Supplementary Figure S5; Zhang et al., 2018b). However, plc7 and plc5/7 mutants were both less sensitive to ABA-induced stomatal closure (Figure 7C and Supplementary Figure S5). Under control conditions, PLC7 is not expressed in guard cells (Figure 1) but levels are strongly upregulated upon ABA treatment (Bauer et al., 2013). Hence, the drought tolerant phenotype in plc5/7 could be a consequence of a local upregulation (e.g., guard cells) of one or more redundant PLCs, which may remain undetectable when whole seedling-mRNA levels are measured (Figure 7D).
Salt- and hyperosmotic stress have been shown to trigger phospholipid-signaling responses in many studies (Hong Y. et al., 2010; Munnik and Zarza, 2013; Hou et al., 2016; Meijer et al., 2017). To mimic the osmotic stress of drought, the effect of sorbitol on 32P-prelabeled seedlings of plc5/7, PLC7-OE and wild type was measured. Under control conditions, no difference in PPIs- and PA levels were found among genotypes, however, in response to sorbitol, a much stronger PIP2 response in both plc5/7 and PLC7-OE lines was found (Figure 9). Whether the above response in plc5/7 is a consequence of enhanced, local expression of another PLC, or a consequence of differential gene expression and hence drought tolerant response, requires further studies. Overexpression of PLC3 and PLC5 also showed an enhanced PIP2 response. Maybe the constitutive hydrolysis of PPIs due to overexpression of PLC, enhances the activity of the lipid kinases to replenish PPI pools, and that hyperosmotic stress activation (sorbitol/drought) causes stronger PIP2 responses and, hence, downstream signaling. Apart from being a precursor of IPPs and PA, PIP2 is also emerging as signaling molecule itself, e.g., involving reorganization of the cytoskeleton, endo- and exocytosis, and ion channel regulation (Xue et al., 2009; Ischebeck et al., 2010; Heilmann and Heilmann, 2015; Heilmann, 2016a,b; Heilmann and Ischebeck, 2016; Gerth et al., 2017; Noack and Jaillais, 2017). Whether PLC performs as a signal generator or PIP2-signaling attenuator remains to be shown and investigated. The identification and characterization of some genuine PIP2 targets will be essential to start unraveling the molecular mechanisms involved.
Author Contributions
RvW, QZ, XZ, ML, AL, and TM designed the experiments. ML, XZ, FM, and WL performed the mucilage experiments, while AG, DS, and CG-M performed the stomatal measurements. RvW, QZ, and ML executed all the remaining experiments. RvW, QZ, AL, MH, and TM wrote the article.
Funding
This work was financially supported by the China Scholarship Council (CSC File No. 201206300058 to QZ), by UNMdP, CONICET and ANPCyT (to AL, CG-M, DS, and AG), and by the Netherlands Organization for Scientific Research (NWO; 867.15.020 to TM).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01721/full#supplementary-material
FIGURE S1 | A plc5/plc7 double mutant is viable and not affected in root architecture. (A) Q-PCR expression levels of PLC5 (left) and PLC7 (right) in wild type and plc5/7-double mutant, using SAND as reference gene. Values are means ± SD (n = 3) of a representative experiment that was repeated three times. (B) Seedling morphology of wild type and plc5/7 (12 DAG). (C) Primary root (PR) length and (D) lateral root (LR) number at 12 DAG. Values are means ± SE of three independent experiments (n > 20). Asterisk (∗) indicate significance at P < 0.05 compared to wild-type, based on Student’s t-test.
FIGURE S2 | Single plc mutants do no show defect in mucilage swelling. Equal amounts of dry seeds (∼1000) of WT, single plc5- and plc7-mutants, and plc5/7 double mutants were immersed in water and photographed after 0 (upper panel) and 12 hrs imbibition (lower panel). Only the plc5/7 double mutants exhibit a mucilage defect and therefor swell less than WT or the individual plc mutants.
FIGURE S3 | Single plc single mutants do not show serration phenotype. Plants were grown for 4 weeks at short-day conditions and photographed. Two separate experiments are shown to show that single mutants look like WT and do not have the serration phenotype as observed for plc5/plc7-double mutants. (A) Plant were grown with either 9 plants/pot (top 3 rows) or 5 plants/pot (lower row). (B) Shows WT, single- and plc5/plc7-double mutants, with the latter clearly showing increased leaf serration.
FIGURE S4 | Sugar composition of wild type- and plc5/7-mutant seeds. Sugars were extracted from dry seeds and quantified by HPAEC-PAD. Quantities were corrected through internal standards, and transformed into mg of sugar per gram of dry material. Values represent the means of triplicates ± SE of three independent experiments.
FIGURE S5 |PLC7-KO and -KD mutants are less sensitive to ABA induced-stomatal closure. Leaves from 3-weeks old plants were stripped and peels were incubated in opening buffer with light for 3 h until stomata were fully open. Peels were then treated with different concentrations of ABA for 90 mins, after which stomata were digitized and the aperture width measured for wild type and plc7-3 (left) or plc7-4 (right). Data were analyzed by one-way ANOVA. For each plant line, treatments were compared against their own control and statistically significant differences between doses are indicated by crosses for wild type or asterisks for transgenic lines (P < 0.05, Dunn’s method). Wild type and different genotypes were compared in control conditions and statically differences are indicated with empty circle (P < 0.001, Mann Whitney). Values are represented by means ± SE of at least three independent experiments (n > 100).
Footnotes
References
Anderson, C. T., Carroll, A., Akhmetova, L., and Somerville, C. (2010). Real-time imaging of cellulose reorientation during cell wall expansion in Arabidopsis roots. Plant Physiol. 152, 787–796. doi: 10.1104/pp.109.150128
Arisz, S. A., Testerink, C., and Munnik, T. (2009). Plant PA signaling via diacylglycerol kinase. Biochim. Biophys. Acta 1791, 869–875. doi: 10.1016/j.bbalip.2009.04.006
Arisz, S. A., van Wijk, R., Roels, W., Zhu, J. K., Haring, M. A., and Munnik, T. (2013). Rapid phosphatidic acid accumulation in response to low temperature stress in Arabidopsis is generated through diacylglycerol kinase. Front. Plant Sci. 4:1. doi: 10.3389/fpls.2013.00001
Balla, T. (2013). Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol. Rev. 93, 1019–1137. doi: 10.1152/physrev.00028.2012
Basu, D., Tian, L., DeBrosse, T., Poirier, E., Emch, K., Herock, H., et al. (2016). Glycosylation of a fasciclin-like arabinogalactan-protein (SOS5) mediates root growth and seed mucilage adherence via a cell wall receptor-like kinase (FEI1/ FEI2) pathway in Arabidopsis. PLoS One 11:e0145092. doi: 10.1371/journal.pone.0145092
Bauer, H., Ache, P., Lautner, S., Fromm, J., Hartung, W., Al-Rasheid, K. A. S., et al. (2013). The stomatal response to reduced relative humidity requires guard cell-autonomous ABA synthesis. Curr. Biol. 23, 53–57. doi: 10.1016/j.cub.2012.11.022
Ben-Tov, D., Idan-Molakandov, A., Hugger, A., Ben-Shlush, I., Günl, M., Yang, B., et al. (2018). The role of COBRA-LIKE 2 function, as part of the complex network of interacting pathways regulating Arabidopsis seed mucilage polysaccharide matrix organization. Plant J. 94, 497–512. doi: 10.1111/tpj.13871
Bilsborough, G. D., Runions, A., Barkoulas, M., Jenkins, H. W., Hasson, A., and Galinha, C. (2011). Model for the regulation of Arabidopsis thaliana leaf margin development. Proc. Natl. Acad. Sci. U.S.A. 108, 3424–3429. doi: 10.1073/pnas.1015162108
Byrne, M. E. (2005). Networks in leaf development. Curr. Opin. Plant Biol. 8, 59–66. doi: 10.1016/j.pbi.2004.11.009
Camoni, L., Di Lucente, C., Pallucca, R., Visconti, S., and Aducci, P. (2012). Binding of phosphatidic acid to 14-3-3 proteins hampers their ability to activate the plant plasma membrane H+-ATPase. IUBMB Life 64, 710–716. doi: 10.1002/iub.1058
Carpita, N. C., and Gibeaut, D. M. (1993). Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J. 3, 1–30. doi: 10.1111/j.1365-313X.1993.tb00007.x
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313X.1998.00343.x
Cosgrove, D. J. (1997). Assembly and enlargement of the primary cell wall in plants. Annu. Rev. Cell Dev. Biol. 13, 171–201. doi: 10.1146/annurev.cellbio.13.1.171
D’Ambrosio, J. M., Couto, D., Fabro, G., Scuffi, D., Lamattina, L., Munnik, T., et al. (2017). Phospholipase C2 Affects MAMP-triggered immunity by modulating ROS production. Plant Physiol. 175, 970–981. doi: 10.1104/pp.17.00173
Di Fino, L. M., D’Ambrosio, J. M., Tejos, R., van Wijk, R., Lamattina, L., Munnik, T., et al. (2017). Arabidopsis phosphatidylinositol-phospholipase C2 (PLC2) is required for female gametogenesis and embryo development. Planta 245, 717–728. doi: 10.1007/s00425-016-2634-z
Distéfano, A. M., Scuffi, D., García-Mata, C., Lamattina, L., and Laxalt, A. M. (2012). Phospholipase Dδ is involved in nitric oxide-induced stomatal closure. Planta 236, 1899–1907. doi: 10.1007/s00425-012-1745-4
Dowd, P. E., Coursol, S., Skirpan, A. L., Kao, T., and Gilroy, S. (2006). Petunia phospholipase C1 is involved in pollen tube growth. Plant Cell 18, 1438–1453. doi: 10.1105/tpc.106.041582.1
Fleming, A. J. (2005). The control of leaf development. New Phytol. 166, 9–20. doi: 10.1111/j.1469-8137.2004.01292.x
Gao, K., Liu, Y. L., Li, B., Zhou, R. G., Sun, D. Y., and Zheng, S. Z. (2014). Arabidopsis thaliana phosphoinositide-specific phospholipase C isoform 3 (AtPLC3) and AtPLC9 have an additive effect on thermotolerance. Plant Cell Physiol. 55, 1873–1883. doi: 10.1093/pcp/pcu116
Gerth, K., Lin, F., Menzel, W., Krishnamoorthy, P., Stenzel, I., Heilmann, M., and Heilmann, I. (2017). Guilt by association: a phenotype-based view of the plant phosphoinositide network. Annu. Rev. Plant Biol. 68, 349–374. doi: 10.1146/annurev-arplant-042916-041022
Georges, F., Das, S., Ray, H., Bock, C., Nokhrina, K., Kolla, V. A., et al. (2009). Over-expression of Brassica napus phosphatidylinositol-phospholipase C2 in canola induces significant changes in gene expression and phytohormone distribution patterns, enhances drought tolerance and promotes early flowering and maturation. Plant, Cell Environ. 32, 1664–1681. doi: 10.1111/j.1365-3040.2009.02027.x
Gillaspy, G. E. (2013). The role of phosphoinositides and inositol phosphates in plant cell signaling. Adv. Exp. Med. Biol. 991, 141–157. doi: 10.1007/978-94-007-6331-9_8
Golz, J. F., Allen, P. J., Li, S. F., Parish, R. W., Jayawardana, N. U., Bacic, A., et al. (2018). Layers of regulation – Insights into the role of transcription factors controlling mucilage production in the Arabidopsis seed coat. Plant Sci. 272, 179–192. doi: 10.1016/j.plantsci.2018.04.021
Griffiths, J. S., and North, H. M. (2017). Sticking to cellulose: exploiting Arabidopsis seed coat mucilage to understand cellulose biosynthesis and cell wall polysaccharide interactions. New Phytol. 214, 959–966. doi: 10.1111/nph.14468
Griffiths, J. S. J., Šola, K., Kushwaha, R., Lam, P., Tateno, M., Young, R., et al. (2015). Unidirectional movement of cellulose synthase complexes in arabidopsis seed coat epidermal cells deposit cellulose involved in mucilage extrusion, adherence, and ray formation. Plant Physiol. 168, 502–520. doi: 10.1104/pp.15.00478
Han, B., Yang, Z., Samma, M. K., Wang, R., and Shen, W. (2013). Systematic validation of candidate reference genes for qRT-PCR normalization under iron deficiency in Arabidopsis. Biometals 26, 403–413. doi: 10.1007/s10534-013-9623-5
Hay, A., Barkoulas, M., and Tsiantis, M. (2006). ASYMMETRIC LEAVES1 and auxin activities converge to repress BREVIPEDICELLUS expression and promote leaf development in Arabidopsis. Development 133, 3955–3961. doi: 10.1242/dev.02545
Hay, A., and Tsiantis, M. (2006). The genetic basis for differences in leaf form between Arabidopsis thaliana and its wild relative Cardamine hirsuta. Nat. Genet. 38, 942–947. doi: 10.1038/ng1835
Heilmann, I. (2016a). Phosphoinositide signaling in plant development. Development 143, 2044–2055. doi: 10.1242/dev.136432
Heilmann, I. (2016b). Plant phosphoinositide signaling - dynamics on demand. Biochim. Biophys. Acta 1861, 1345–1351. doi: 10.1016/j.bbalip.2016.02.013
Heilmann, I., and Ischebeck, T. (2016). Male functions and malfunctions: the impact of phosphoinositides on pollen development and pollen tube growth. Plant Reprod. 29, 3–20. doi: 10.1007/s00497-015-0270-6
Heilmann, M., and Heilmann, I. (2015). Plant phosphoinositides - complex networks controlling growth and adaptation. Biochim. Biophys. Acta 1851, 759–769. doi: 10.1016/j.bbalip.2014.09.018
Helling, D., Possart, A., Cottier, S., Klahre, U., and Kost, B. (2006). Pollen tube tip growth depends on plasma membrane polarization mediated by tobacco PLC3 activity and endocytic membrane recycling. Plant Cell 18, 3519–3534. doi: 10.1105/tpc.106.047373
Hite, R. K., Butterwick, J. A., and MacKinnon, R. (2014). Phosphatidic acid modulation of Kv channel voltage sensor function. eLife 3:e04366. doi: 10.7554/eLife.04366
Hong, S. M., Bahn, S. C., Lyu, A., Jung, H. S., and Ahn, J. H. (2010). Identification and testing of superior reference genes for a starting pool of transcript normalization in Arabidopsis. Plant Cell Physiol. 51, 1694–1706. doi: 10.1093/pcp/pcq128
Hong, Y., Zhang, W., and Wang, X. (2010). Phospholipase D and phosphatidic acid signalling in plant response to drought and salinity. Plant Cell Environ. 33, 627–635. doi: 10.1111/j.1365-3040.2009.02087.x
Hou, Q., Ufer, G., and Bartels, D. (2016). Lipid signalling in plant responses to abiotic stress. Plant, Cell Environ. 39, 1029–1048. doi: 10.1111/pce.12666
Hu, R. (2016). Irregular xylem 7 (IRX7) is required for anchoring seed coat mucilage in Arabidopsis. Plant Mol. Biol. 92, 25–38. doi: 10.1007/s11103-016-0493-4
Hu, R., Li, J., Wang, X., Zhao, X., Yang, X., Tang, Q., et al. (2016). Xylan synthesized by irregular xylem 14 (IRX14) maintains the structure of seed coat mucilage in Arabidopsis. J. Exp. Bot. 67, 1243–1257. doi: 10.1093/jxb/erv510
Hunt, L., Mills, L. N., Pical, C., Leckie, C. P., Aitken, F. L., Kopka, J., et al. (2003). Phospholipase C is required for the control of stomatal aperture by ABA. Plant J. 34, 47–55. doi: 10.1046/j.1365-313X.2003.01698.x
Hunt, L., Otterhag, L., Lee, J. C., Lasheen, T., Hunt, J., Seki, M., et al. (2004). Gene-specific expression and calcium activation of Arabidopsis thaliana phospholipase C isoforms. New Phytol. 162, 643–654. doi: 10.1111/j.1469-8137.2004.01069.x
Irvine, R. F. (2006). Nuclear inositide signalling-expansion, structures and clarification. Biochim. Biophys. Acta 1761, 505–508. doi: 10.1016/j.bbalip.2006.02.008.Nuclear
Ischebeck, T., Seiler, S., and Heilmann, I. (2010). At the poles across kingdoms: phosphoinositides and polar tip growth. Protoplasma 240, 13–31. doi: 10.1007/s00709-009-0093-0
Julkowska, M. M., McLoughlin, F., Galvan-Ampudia, C. S., Rankenberg, J. M., Kawa, D., Klimecka, M., et al. (2015). Identification and functional characterization of the Arabidopsis Snf1-related protein kinase SnRK2.4 phosphatidic acid-binding domain. Plant. Cell Environ. 38, 614–624. doi: 10.1111/pce.12421
Kawamura, E., Horiguchi, G., and Tsukaya, H. (2010). Mechanisms of leaf tooth formation in Arabidopsis. Plant J. 62, 429–441. doi: 10.1111/j.1365-313X.2010.04156.x
Kuo, H. F., Chang, T. Y., Chiang, S. F., Wang, W., Di Charng, Y. Y., and Chiou, T. J. (2014). Arabidopsis inositol pentakisphosphate 2-kinase, AtIPK1, is required for growth and modulates phosphate homeostasis at the transcriptional level. Plant J. 80, 503–515. doi: 10.1111/tpj.12650
Kuo, H.-F., Hsu, Y.-Y., Lin, W.-C., Chen, K.-Y., Munnik, T., Brearley, C. A., et al. (2018). Arabidopsis inositol phosphate kinases IPK1 and ITPK1 constitute a metabolic pathway in maintaining phosphate homeostasis. Plant J. 95, 613–630. doi: 10.1111/tpj.13974
Laha, D., Johnen, P., Azevedo, C., Dynowski, M., Weiß, M., Capolicchio, S., et al. (2015). VIH2 regulates the synthesis of inositol pyrophosphate InsP8 and jasmonate-dependent defenses in Arabidopsis. Plant Cell 27, 1082–1097. doi: 10.1105/tpc.114.135160
Laha, D., Parvin, N., Dynowski, M., Johnen, P., Mao, H., Bitters, S. T., et al. (2016). Inositol polyphosphate binding specificity of the jasmonate receptor complex. Plant Physiol. 171, 2364–2370. doi: 10.1104/pp.16.00694
Lee, H. S., Lee, D. H., Cho, H. K., Kim, S. H., Auh, J. H., and Pai, H. S. (2015). InsP6-sensitive variants of the Gle1 mRNA export factor rescue growth and fertility defects of the ipk1 low-phytic-acid mutation in Arabidopsis. Plant Cell 27, 417–431. doi: 10.1105/tpc.114.132134
Lemtiri-Chlieh, F., MacRobbie, E. A. C., and Brearley, C. A. (2000). Inositol hexakisphosphate is a physiological signal regulating the K+-inward rectifying conductance in guard cells. Proc. Natl. Acad. Sci. U.S.A. 97, 8687–8692. doi: 10.1073/pnas.140217497
Lemtiri-Chlieh, F., MacRobbie, E. A. C., Webb, A. A. R., Manison, N. F., Brownlee, C., Skepper, J. N., et al. (2003). Inositol hexakisphosphate mobilizes an endomembrane store of calcium in guard cells. Proc. Natl. Acad. Sci. U.S.A. 100, 10091–10095. doi: 10.1073/pnas.1133289100
Li, J., Pleskot, R., Henty-Ridilla, J. L., Blanchoin, L., Potockı, M., and Staiger, C. J. (2012). Arabidopsis capping protein senses cellular phosphatidic acid levels and transduces these into changes in actin cytoskeleton dynamics. Plant Signal. Behav. 7, 1727–1730. doi: 10.4161/psb.22472
Li, L., He, Y., Wang, Y., Zhao, S., Chen, X., Ye, T., et al. (2015). Arabidopsis PLC2 is involved in auxin-modulated reproductive development. Plant J. 84, 504–515. doi: 10.1111/tpj.13016
Li, M., Hong, Y., and Wang, X. (2009). Phospholipase D- and phosphatidic acid-mediated signaling in plants. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1791, 927–935. doi: 10.1016/j.bbalip.2009.02.017
Macquet, A., Ralet, M. C., Kronenberger, J., Marion-Poll, A., and North, H. M. (2007). In situ, chemical and macromolecular study of the composition of Arabidopsis thaliana seed coat mucilage. Plant Cell Physiol. 48, 984–999. doi: 10.1093/pcp/pcm068
McLoughlin, F., Galvan-Ampudia, C. S., Julkowska, M. M., Caarls, L., van der Does, D., Lauriere, C., et al. (2012). The Snf1-related protein kinases SnRK2.4 and SnRK2.10 are involved in maintenance of root system architecture during salt stress. Plant J. 72, 436–449. doi: 10.1111/j.1365-313X.2012.05089.x
McLoughlin, F., and Testerink, C. (2013). Phosphatidic acid, a versatile water-stress signal in roots. Front. Plant Sci. 23:525. doi: 10.3389/fpls.2013.00525
Meijer, H. J. G., van Himbergen, J. A. J., Musgrave, A., and Munnik, T. (2017). Acclimation to salt modifies the activation of several osmotic stress-activated lipid signalling pathways in Chlamydomonas. Phytochemistry 135, 64–72. doi: 10.1016/j.phytochem.2016.12.014
Mendu, V., Griffiths, J. S., Persson, S., Stork, J., Downie, B., Voiniciuc, C., et al. (2011). Subfunctionalization of cellulose synthases in seed coat epidermal cells mediates secondary radial wall synthesis and mucilage attachment. Plant Physiol. 157, 441–453. doi: 10.1104/pp.111.179069
Michell, R. H. (2008). Inositol derivatives: evolution and functions. Nat. Rev. 9, 151–161. doi: 10.1038/nrm2334
Mishra, G., Zhang, W., Deng, F., Zhao, J., and Wang, X. (2006). A bifurcating pathway directs abscisic acid effects on stomatal closure and opening in Arabidopsis. Science 312, 264–266. doi: 10.1126/science.1123769
Mills, L. N., Hunt, L., Leckie, C. P., Aitken, F. L., Wentworth, M., McAinsh, M. R., et al. (2004). The effects of manipulating phospholipase C on guard cell ABA-signalling. J. Exp. Bot. 55, 199–204.
Monserrate, J. P., and York, J. D. (2010). Inositol phosphate synthesis and the nuclear processes they affect. Curr. Opin. Cell Biol. 22, 365–373. doi: 10.1016/j.ceb.2010.03.006
Mosblech, A., König, S., Stenzel, I., Grzeganek, P., Feussner, I., and Heilmann, I. (2008). Phosphoinositide and inositolpolyphosphate signalling in defense responses of Arabidopsis thaliana challenged by mechanical wounding. Mol. Plant 1, 249–261. doi: 10.1093/mp/ssm028
Mosblech, A., Thurow, C., Gatz, C., Feussner, I., and Heilmann, I. (2011). Jasmonic acid perception by COI1 involves inositol polyphosphates in Arabidopsis thaliana. Plant J. 65, 949–957. doi: 10.1111/j.1365-313X.2011.04480.x
Munnik, T. (2001). Phosphatidic acid: an emerging plant lipid second messenger. Trends Plant Sci. 6, 227–233. doi: 10.1016/S1360-1385(01)01918-5
Munnik, T. (2014). “PI-PLC: phosphoinositide-phospholipase C in plant signalling,” in Phospholipases in Plant Signaling, ed. X. Wang (Berlin: Springer-Verlag), 27–54.
Munnik, T., and Nielsen, E. (2011). Green light for polyphosphoinositide signals in plants. Curr. Opin. Plant Biol. 14, 489–497. doi: 10.1016/j.pbi.2011.06.007
Munnik, T., and Testerink, C. (2009). Plant phospholipid signaling: “in a nutshell”. J. Lipid Res. 50, S260–S265. doi: 10.1194/jlr.R800098-JLR200
Munnik, T., and Vermeer, J. E. M. (2010). Osmotic stress-induced phosphoinositide and inositol phosphate signalling in plants. Plant. Cell Environ. 33, 655–669. doi: 10.1111/j.1365-3040.2009.02097.x
Munnik, T., and Zarza, X. (2013). Analyzing plant signaling phospholipids through 32Pi-labeling and TLC. Methods Mol. Biol. 1009, 3–15. doi: 10.1007/978-1-62703-401-2_1
Murphy, A. M., Otto, B., Brearley, C. A., Carr, J. P., and Hanke, D. E. (2008). A role for inositol hexakisphosphate in the maintenance of basal resistance to plant pathogens. Plant J. 56, 638–652. doi: 10.1111/j.1365-313X.2008.03629.x
Nikovics, K., Blein, T., Peaucelle, A., Ishida, T., Morin, H., Aida, M., et al. (2006). The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell 18, 2929–2945. doi: 10.1105/tpc.106.045617
Noack, L. C., and Jaillais, Y. (2017). Precision targeting by phosphoinositides: how PIs direct endomembrane trafficking in plants. Curr. Opin. Plant Biol. 40, 22–33. doi: 10.1016/j.pbi.2017.06.017
Pleskot, R., Li, J., Žárskı, V., Potockı, M., and Staiger, C. J. (2017). Regulation of cytoskeletal dynamics by phospholipase D and phosphatidic acid. Trends Plant Sci. 18, 496–504. doi: 10.1016/j.tplants.2013.04.005
Pleskot, R., Potockı, M., Pejchar, P., Linek, J., Bezvoda, R., Martinec, J., et al. (2010). Mutual regulation of plant phospholipase D and the actin cytoskeleton. Plant J. 62, 494–507. doi: 10.1111/j.1365-313X.2010.04168.x
Pokotylo, I., Kolesnikov, Y., Kravets, V., Zachowski, A., and Ruelland, E. (2014). Plant phosphoinositide-dependent phospholipases C: variations around a canonical theme. Biochimie 96, 144–157. doi: 10.1016/j.biochi.2013.07.004
Pokotylo, I., Kravets, V., Martinec, J., and Ruelland, E. (2018). The phosphatidic acid paradox: too many actions for one molecule class? Lessons from plants. Prog. Lipid Res. 71, 43–53. doi: 10.1016/j.plipres.2018.05.003
Pokotylo, I., Pejchar, P., Potocky, M., Kocourkova, D., Krckova, Z., Ruelland, E., et al. (2013). The plant non-specific phospholipase C gene family. Novel competitors in lipid signalling. Prog. Lipid Res. 52, 62–79. doi: 10.1016/j.plipres.2012.09.001
Puga, M. I., Mateos, I., Charukesi, R., Wang, Z., Franco-Zorrilla, J. M., de Lorenzo, L., et al. (2014). SPX1 is a phosphate-dependent inhibitor of phosphate starvation response 1 in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 111, 14947–14952. doi: 10.1073/pnas.1404654111
Ralet, M.-C. C., Crepeau, M. J., Vigouroux, J., Tran, J., Berger, A., Salle, C., et al. (2016). Xylans provide the structural driving force for mucilage adhesion to the arabidopsis seed coat. Plant Physiol. 171, 165–178. doi: 10.1104/pp.16.00211
Repp, A., Mikami, K., Mittmann, F., and Hartmann, E. (2004). Phosphoinositide-specific phospholipase C is involved in cytokinin and gravity responses in the moss Physcomitrella patens. Plant J. 40, 250–259. doi: 10.1111/j.1365-313X.2004.02205.x
Ribeiro, P. R., Fernandez, L. G., de Castro, R. D., Ligterink, W., and Hilhorst, H. W. (2014). Physiological and biochemical responses of Ricinus communis seedlings to different temperatures: a metabolomics approach. BMC Plant Biol. 14:223. doi: 10.1186/s12870-014-0223-5
Saez-Aguayo, S., Rondeau-Mouro, C., Macquet, A., Kronholm, I., Ralet, M. C., Berger, A., et al. (2014). Local evolution of seed flotation in Arabidopsis. PLoS Genet. 10:e1004221. doi: 10.1371/journal.pgen.1004221
Sanchez, J. P., and Chua, N. H. (2001). Arabidopsis PLC1 is required for secondary responses to abscisic acid signals. Plant Cell 13, 1143–1154. doi: 10.1105/tpc.13.5.1143
Scarpella, E., Marcos, D., Friml, J., Berleth, T., Scarpella, E., Marcos, D., et al. (2006). Control of leaf vascular patterning by polar auxin transport control of leaf vascular patterning by polar auxin transport. Genes Dev. 20, 1015–1027. doi: 10.1101/gad.1402406
Sean, R., Pedro, L., Ruth, R., and Suzanne, R. (2010). Abscisic acid:emergence of a core signaling network. Annu. Rev. Plant Biol. 61, 651–679. doi: 10.1146/annurev-arplant-042809-112122
Sheard, L. B., Tan, X., Mao, H., Withers, J., Ben-Nissan, G., Hinds, T. R., et al. (2010). Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468, 400–405. doi: 10.1038/nature09430
Sullivan, S., Ralet, M. M.-C., Berger, A., Diatloff, E., Bischoff, V., Gonneau, M., et al. (2011). CESA5 is required for the synthesis of cellulose with a role in structuring the adherent mucilage of Arabidopsis seeds. Plant Physiol. 156, 1725–1739. doi: 10.1104/pp.111.179077
Tan, X., Calderon-Villalobos, L. I. A., Sharon, M., Zheng, C., Robinson, C. V., Estelle, M., et al. (2007). Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446, 640–645. doi: 10.1038/nature05731
Tasma, I. M., Brendel, V., Whitham, S. A., and Bhattacharyya, M. K. (2008). Expression and evolution of the phosphoinositide-specific phospholipase C gene family in Arabidopsis thaliana. Plant Physiol. Biochem. 46, 627–637. doi: 10.1016/j.plaphy.2008.04.015
Testerink, C., Dekker, H. L., Lim, Z.-Y., Johns, M. K., Holmes, A. B., Koster, C. G., et al. (2004). Isolation and identification of phosphatidic acid targets from plants. Plant J. 39, 527–536. doi: 10.1111/j.1365-313X.2004.02152.x
Testerink, C., and Munnik, T. (2011). Molecular, cellular, and physiological responses to phosphatidic acid formation in plants. J. Exp. Bot. 62, 2349–2361. doi: 10.1093/jxb/err079
Thomas, C., and Staiger, C. J. (2014). A dynamic interplay between membranes and the cytoskeleton critical for cell development and signaling. Front. Plant Sci. 5:335. doi: 10.3389/fpls.2014.00335
Thota, S. G., and Bhandari, R. (2015). The emerging roles of inositol pyrophosphates in eukaryotic cell physiology. J. Biosci. 40, 593–605. doi: 10.1007/s12038-015-9549-x
Tripathy, M. K., Tyagi, W., Goswami, M., Kaul, T., Singla-Pareek, S. L., Deswal, R., et al. (2011). Characterization and functional validation of tobacco PLC delta for abiotic stress tolerance. Plant Mol. Biol. Rep. 30, 488–497. doi: 10.1007/s11105-011-0360-z
Tsukaya, H. (2006). Mechanism of leaf-shape determination. Annu. Rev. Plant Biol. 57, 477–496. doi: 10.1146/annurev.arplant.57.032905.105320
Tsukaya, H., and Uchimiya, H. (1997). Genetic analyses of the formation of the serrated margin of leaf blades in Arabidopsis: combination of a mutational analysis of leaf morphogenesis with the characterization of a specific marker gene expressed in hydathodes and stipules. Mol. Gen. Genet. 256, 231–238. doi: 10.1007/s004380050565
Ufer, G., Gertzmann, A., Gasulla, F., Rohrig, H., and Bartels, D. (2017). Identification and characterization of the phosphatidic acid-binding A. thaliana phosphoprotein PLDrp1 that is regulated by PLDα1 in a stress-dependent manner. Plant J. 92, 276–290. doi: 10.1111/tpj.13651
van Leeuwen, W., Vermeer, J. E. M., Gadella, T. W. J., and Munnik, T. (2007). Visualization of phosphatidylinositol 4,5-bisphosphate in the plasma membrane of suspension-cultured tobacco BY-2 cells and whole Arabidopsis seedlings. Plant J. 52, 1014–1026. doi: 10.1111/j.1365-313X.2007.03292.x
Voiniciuc, C., Günl, M., Schmidt, M. H.-W., and Usadel, B. (2015a). Highly branched Xylan made by IRREGULAR XYLEM14 and MUCILAGE-RELATED21 links mucilage to Arabidopsis seeds. Plant Physiol. 169, 2481–2495. doi: 10.1104/pp.15.01441
Voiniciuc, C., Yang, B., Schmidt, M. H.-W., Günl, M., and Usadel, B. (2015b). Starting to gel: how Arabidopsis seed coat epidermal cells produce specialized secondary cell walls. Int. J. Mol. Sci. 16, 3452–3473. doi: 10.3390/ijms16023452
Vossen, J. H., Abd-El-Haliem, A., Fradin, E. F., Van Den Berg, G. C. M., Ekengren, S. K., Meijer, H. J. G., et al. (2010). Identification of tomato phosphatidylinositol-specific phospholipase-C (PI-PLC) family members and the role of PLC4 and PLC6 in HR and disease resistance. Plant J. 62, 224–239. doi: 10.1111/j.1365-313X.2010.04136.x
Wallace, I. S., and Anderson, C. T. (2012). Small molecule probes for plant cell wall polysaccharide imaging. Front. Plant Sci. 3:89. doi: 10.3389/fpls.2012.00089
Wang, C. R., Yang, A. F., Yue, G. D., Gao, Q., Yin, H. Y., and Zhang, J. R. (2008). Enhanced expression of phospholipase C 1 (ZmPLC1) improves drought tolerance in transgenic maize. Planta 227, 1127–1140. doi: 10.1007/s00425-007-0686-9
Wang, X., Devaiah, S. P., Zhang, W., and Welti, R. (2006). Signaling functions of phosphatidic acid. Prog. Lipid Res. 45, 250–278. doi: 10.1016/j.plipres.2006.01.005
Western, T. L., Skinner, D. J., and Haughn, G. W. (2000). Differentiation of mucilage secretory cells of the Arabidopsis seed coat. Plant Physiol. 122, 345–356. doi: 10.1104/pp.122.2.345
Wild, R., Gerasimaite, R., Jung, J., Truffault, V., Pavlovic, I., Schmidt, A., et al. (2016). Control of eukaryotic phosphate homeostasis by inositol polyphosphate sensor domains. Science 352, 986–990. doi: 10.1126/science.aad9858
Williams, S. P., Gillaspy, G. E., and Perera, I. Y. (2015). Biosynthesis and possible functions of inositol pyrophosphates in plants. Front. Plant Sci. 6:67. doi: 10.3389/fpls.2015.00067
Winter, D., Vinegar, B., Nahal, H., Ammar, R., Wilson, G. V., and Provart, N. J. (2007). An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2:e718. doi: 10.1371/journal.pone.0000718
Xue, H. W., Chen, X., and Mei, Y. (2009). Function and regulation of phospholipid signalling in plants. Biochem. J. 421, 145–156. doi: 10.1042/BJ20090300
Yao, H., and Xue, H. (2018). Phosphatidic acid plays key roles regulating plant development and stress responses. J. Integr. Plant Biol. 60, 851–863. doi: 10.1111/jipb.12655
Zhang, Q., van Wijk, R., Shahbaz, M., Roels, W., Schooten, B., van Vermeer, J. E. M., et al. (2018a). Arabidopsis phospholipase C3 is involved in lateral root initiation and ABA responses in seed germination and stomatal closure. Plant Cell Physiol. 59, 469–486. doi: 10.1093/pcp/pcx194
Zhang, Q., van Wijk, R., Zarza, X., Shahbaz, M., van Hooren, M., Guardia, A., et al. (2018b). Knock-down of Arabidopsis PLC5 reduces primary root growth and secondary root formation while overexpression improves drought tolerance and causes stunted root hair growth. Plant Cell Physiol. 59, 2004–2019. doi: 10.1093/pcp/pcy120
Zhao, X., Qiao, L., and Wu, A. (2017). Effective extraction of Arabidopsis adherent seed mucilage by ultrasonic treatment. Sci. Rep. 7:40672. doi: 10.1038/srep40672
Keywords: PLC, seed mucilage, leaf serration, ABA sensitivity, drought tolerance
Citation: van Wijk R, Zhang Q, Zarza X, Lamers M, Marquez FR, Guardia A, Scuffi D, García-Mata C, Ligterink W, Haring MA, Laxalt AM and Munnik T (2018) Role for Arabidopsis PLC7 in Stomatal Movement, Seed Mucilage Attachment, and Leaf Serration. Front. Plant Sci. 9:1721. doi: 10.3389/fpls.2018.01721
Received: 24 September 2018; Accepted: 05 November 2018;
Published: 27 November 2018.
Edited by:
Yvon Jaillais, École Normale Supérieure de Lyon, FranceReviewed by:
Eric Ruelland, Centre National de la Recherche Scientifique (CNRS), FranceHenri Batoko, Université catholique de Louvain, Belgium
Catalin Voiniciuc, Heinrich Heine Universität Düsseldorf, Germany
Copyright © 2018 van Wijk, Zhang, Zarza, Lamers, Marquez, Guardia, Scuffi, García-Mata, Ligterink, Haring, Laxalt and Munnik. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Teun Munnik, t.munnik@uva.nl
†Present address: Mart Lamers, Department of Viroscience, Erasmus MC, Rotterdam, Netherlands; Wilco Ligterink, Keygene, Wageningen, Netherlands
 Ringo van Wijk
Ringo van Wijk Qianqian Zhang
Qianqian Zhang Xavier Zarza
Xavier Zarza Mart Lamers1†
Mart Lamers1† Francisca Reyes Marquez
Francisca Reyes Marquez Carlos García-Mata
Carlos García-Mata Wilco Ligterink
Wilco Ligterink Michel A. Haring
Michel A. Haring Teun Munnik
Teun Munnik