- Plant Breeding, Wageningen University & Research, Wageningen, Netherlands
In the field, plants constantly face a plethora of abiotic and biotic stresses that can impart detrimental effects on plants. In response to multiple stresses, plants can rapidly reprogram their transcriptome through a tightly regulated and highly dynamic regulatory network where WRKY transcription factors can act as activators or repressors. WRKY transcription factors have diverse biological functions in plants, but most notably are key players in plant responses to biotic and abiotic stresses. In tomato there are 83 WRKY genes identified. Here we review recent progress on functions of these tomato WRKY genes and their homologs in other plant species, such as Arabidopsis and rice, with a special focus on their involvement in responses to abiotic and biotic stresses. In particular, we highlight WRKY genes that play a role in plant responses to a combination of abiotic and biotic stresses.
Introduction
WRKY transcription factors (WRKYs) are a large family of transcriptional regulators, which are defined by the highly conserved WRKY domain (the WRKYGQK motif at the end of the N-terminal and a zinc-finger-like motif at the C-terminus) (Rushton et al., 2010). WRKYs are categorized into three groups (Rushton et al., 2010; Rinerson et al., 2015). Group I (with two WRKY domains) and Group II (with one WRKY domain) contain the zinc-finger-like motif C2–H2 (C–X4-5–C–X22-23–H–X1–H). Group III contains one WRKY domain and a C2–HC zinc-finger-like motif (C–X7–C–X23–H–X1–C) (Eulgem et al., 2000). Based on the primary amino acid sequences, Group II can be further divided into three subgroups (Zhang and Wang, 2005).
Through the binding of the WRKY domain to the W-box cis-acting element (consensus sequence: (T)(T)TGAC(C/T)) in the promoters of their target genes, WRKYs can act as transcriptional activators or repressors in regulatory cascades (Rushton et al., 2010; Yokotani et al., 2013; Bakshi and Oelmuller, 2014). The functional specificity of WRKYs is defined by many factors including the W-box (Yan et al., 2013), the WRKY domain (Cheng et al., 2015), interactions with other proteins (Brand et al., 2013; Franco-Zorrilla et al., 2014), and post-translational modifications (Lai et al., 2011).
Many WRKYs have been identified in the plant kingdom (Supplementary Table S1). Numerous expression and functional studies have given insight in the involvement of WRKYs in different aspects of plant biology (Van Esse et al., 2009; Rushton et al., 2010; Ishihama and Yoshioka, 2012; Hu et al., 2013; Bakshi and Oelmuller, 2014; Yang et al., 2016). Tomato (Solanum lycopersicum) has 83 SlWRKY genes (Huang et al., 2012; Karkute et al., 2018). This review focuses on tomato SlWRKY genes with regard to their roles in plant responses to biotic and abiotic stresses. The nomenclature of the SlWRKY genes follows that of Huang et al. (2012) and Karkute et al. (2018). For SlWRKY genes that have not been studied in detail yet, we propose potential roles in response to (a)biotic stresses by looking at their homologs in other plant species (Supplementary Figure S1). We paid special attention to the role of WRKY genes in the complex regulatory process of plant responses to combined stresses.
Biotic Stress-Related WRKYs
Plants have developed two layers of induced defense responses (Jones and Dangl, 2006), in which WRKYs are shown to function as either positive or negative regulators (e.g., Bakshi and Oelmuller, 2014; Sarris et al., 2015). The first layer, termed PAMP-triggered immunity (PTI), is activated by the recognition between pathogen-associated molecular patterns (PAMPs) and plant’s pattern recognition receptors. Adapted pathogens can express effector proteins to suppress PTI. The second layer [named effector-triggered immunity (ETI)] is triggered by the recognition of pathogen effectors by plant resistance (R) proteins. Plant R proteins usually comprise nucleotide binding-leucine rich repeat (NB-LRR). PTI and ETI induce both local and systemic acquired resistance responses through the production of reactive oxygen species (ROS) and activation of an integrated signaling network including MAP kinases and hormonal signaling pathways (Dodds and Rathjen, 2010). Salicylic acid (SA), jasmonic acid (JA) and ethylene (ET) are the classical immunity-related hormones.
WRKYs are involved in PTI and ETI at different regulatory levels (Bakshi and Oelmuller, 2014). Firstly, WRKYs can interact (in)directly with PAMPs or effector proteins to activate or repress both PTI and ETI. In barley (Hordeum vulgare), HvWRKY1 and HvWRKY2 were activated by flg22 (a MAMP) and acted as repressors of PTI against the powdery mildew fungus Blumeria graminis f.sp. hordei. In addition, the fungal effector AVRA10 activated a specific association between the R protein MLA10 and HvWRKY1/2 leading to inactivation of the repressor function of HvWRKY1/2 (Shen et al., 2007). In Arabidopsis, AtWRKY18, AtWRKY40, and AtWRKY60, homologs of HvWRKY1 and HvWRKY2 (Shen et al., 2007), showed redundant function in negatively regulating PTI to Pseudomonas syringae (Xu et al., 2006) and the powdery mildew fungus Golovinomyces orontii (Shen et al., 2007). Activation of defense-related genes was observed in wrky18 wrky40 and wrky18 wrky60 double mutants and the wrky18 wrky40 wrky60 triple mutants (Xu et al., 2006; Shen et al., 2007). Similarly, the rice (Oryza sativa) OsWRKY62 gene functions as a negative regulator of both PTI and ETI (conferred by the Xa21 gene) to Xanthomonas oryzae (Peng et al., 2008). These WRKYs are members of the WRKY II-a subfamily and the results above suggest that members of this subfamily may have a conserved negative regulatory function in plant defense. However, overexpression of the WRKY II-a subfamily member OsWRKY71 enhanced resistance to Xoo in rice (Liu et al., 2007). Secondly, WRKYs can be regulated by mitogen-activated protein kinases (MAPKs) (Pandey and Somssich, 2009; Ishihama and Yoshioka, 2012). In Nicotiana benthamiana, NtWRKY7, NtWRKY8, NtWRKY9, and NtWRKY11, phosphorylated by pathogen-responsive MAPKs, were able to bind to the W-box in the promoter of the RBOHB gene leading to ROS burst (Ishihama and Yoshioka, 2012; Adachi et al., 2015). AtWRKY33 interacted with MPK4 and MAP kinase 4 substrate 1 (MKS1) (Andreasson et al., 2005). Upon being challenged with P. syringae or upon elicitation by the MAMP flg22, AtWRKY33 was released from this trimeric complex and subsequently bound to the promotor region of Phytoalexin Deficient3 (PAD3) facilitating the synthesis of antimicrobial camalexin (Qiu et al., 2008; Mao et al., 2011; Ishihama and Yoshioka, 2012). Thirdly, WRKYs regulate hormonal signaling pathways. For example, overexpression of AtWRKY18 and AtWRKY70 led to induced expression of defense-related genes, including SA-induced PR1 (Li et al., 2004). The increased susceptibility to Botrytis cinerea of the atwrky33 Arabidopsis mutant was associated with SA-mediated repression of the JA pathway (Birkenbihl et al., 2012). In addition, WRKYs can contribute to plant immunity by modulating small RNAs (smRNAs), by epigenetic mechanisms through histone methylation, as well as by proteasome-mediated degradation and inter-organelle retrograde signaling (Bakshi and Oelmuller, 2014; Phukan et al., 2016).
In tomato, WRKYs are studied for their roles in plant defense by either overexpression and/or silencing them (Supplementary Table S2 and Figures 1, 2). Many tomato WRKYs function as positive regulators of plant responses to biotic stresses. SlWRKY31 (named SlDRW1 in Liu et al., 2014) and SlWRKY33 (named SlWRKY33B and SlWRKY33A in Zhou et al., 2015), homologs of AtWRKY33, were able to complement the compromised tolerance to B. cinerea of the atwrky33 mutant (Zheng et al., 2006). Additionally, overexpression of the Solanum pimpinellifolium allele of SlWRKY33 (named SpWRKY1 in Li et al., 2015a,b) resulted in resistance to the hemi-biotrophic oomycetes Phytophthora nicotianae in tobacco and Phytophthora infestans in tomato. The SlWRKY39 gene, homolog of AtWRKY40, was significantly upregulated in tomato upon being challenged with P. syringae (Huang et al., 2012) and tomato lines over-expressing SlWRKY39 showed enhanced resistance to this pathogen (Sun et al., 2015). Overexpression of SlWRKY45, another homolog of AtWRKY40, enhanced tomato susceptibility to the root-knot nematode Meloidogyne javanica, which was associated with decreased expression of JA- and SA marker genes (Chinnapandi et al., 2017). SlWRKY72, SlWRKY73, or SlWRKY74 (SlWRKY72a or SlWRKY72b in Bhattarai et al., 2010) contributed positively to both PTI and Mi-1-mediated ETI against root-knot nematodes (M. javanica) and potato aphids (Macrosiphum euphorbiae) (Bhattarai et al., 2010). Also, SlWRKY80 (SlWRKY70 in Atamian et al., 2012) was required for Mi-1-mediated resistance against potato aphids and nematodes.
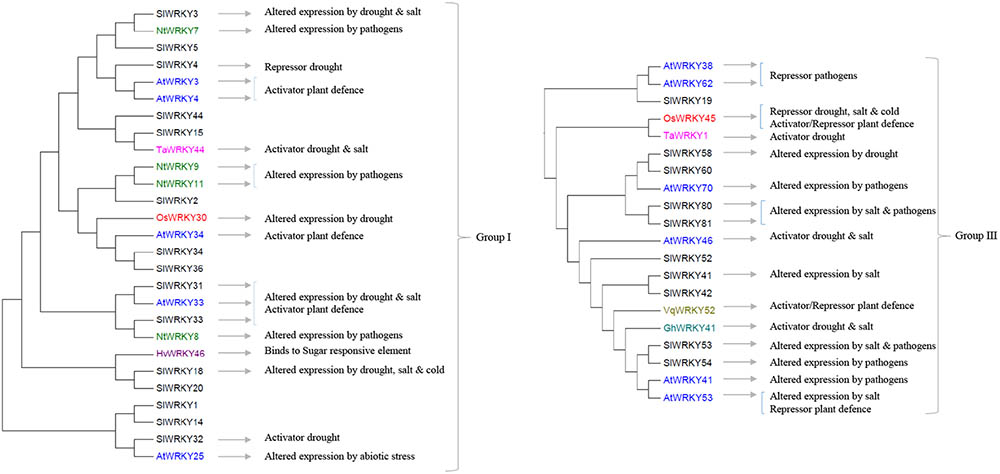
FIGURE 1. The involvements of Group I and III tomato SlWRKY genes and their homologs (highlighted in different colors) in plant responses to biotic and abiotic stresses. The phylogenetic relations of tomato SlWRKYs and their homologs in Arabidopsis (AtWRKYs), rice (OsWRKYs), tobacco (NtWRKY), wheat (TaWRKY), barley (HvWRKY), cotton (GhWRKY), and grape (VqWKRY) are based on the phylogenetic tree presented in Supplementary Figure S1.
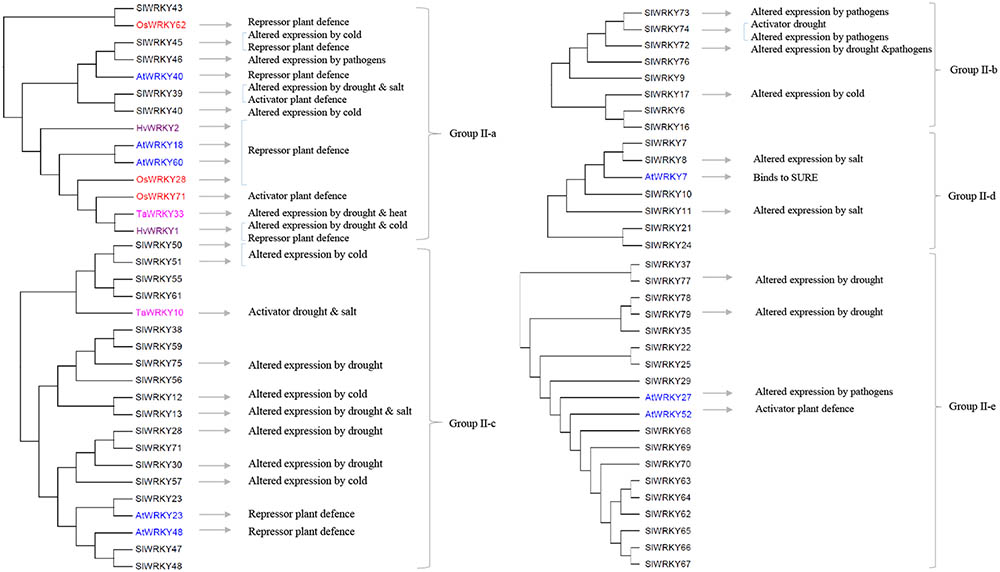
FIGURE 2. The involvements of the Group II tomato SlWRKY genes and their homologs (highlighted in different colors) in plant responses to biotic and abiotic stresses. The phylogenetic relations of tomato SlWRKYs and their homologs in Arabidopsis (AtWRKYs), rice (OsWRKYs), wheat (TaWRKY), and barley (HvWRKY) are based on the phylogenetic tree presented in Supplementary Figure S1.
Upon infection of pathogens, altered expression was reported for several tomato WRKYs, including SlWRKY23 (homolog of AtWRKY23), SlWRKY46 (homolog of AtWRKY40), SlWRKY53/54 (homolog of AtWRKY23), SlWRKY80 and SlWRKY81 (homologs of AtWRKY38 and AtWRKY62) (Huang et al., 2012, 2016; Du et al., 2015; Lucioli et al., 2016; Rezzonico et al., 2017). Their homologs in Arabidopsis act as negative regulators of plant defense: AtWRKY38, AtWRKY48, and AtWRKY62 in the response to P. syringae (Xu et al., 2006; Kim et al., 2008; Xing et al., 2008), AtWRKY23 in response to the nematode Heterodera schachtii (Grunewald et al., 2008), and AtWRKY27 and AtWRKY53 in response to Ralstonia solanacearum (Murray et al., 2007; Mukhtar et al., 2008). Interestingly, overexpression of the grape (Vitis quinquangularis) VqWRKY52 gene in Arabidopsis, a homolog of AtWRKY53 and SlWRKY53/54, enhanced resistance to Golovinomyces cichoracearum and P. syringae, but increased susceptibility to B. cinerea, which was associated with increased expression of SA-pathway related genes and enhanced cell death (Wang et al., 2017). Therefore, further functional analysis of these tomato WRKY genes is needed to confirm their role in either enhanced resistance or increased susceptibility to certain pathogens.
Abiotic Stress-Related WRKYs
A number of studies demonstrate that WRKYs are involved in plant responses to abiotic stresses, such as drought and salinity (Supplementary Table S2 and Figures 1, 2). Expression of genes responsive to the signaling hormone ABA was altered in AtWRKY40 and AtWRKY40/AtWRKY18 knockout lines. Overexpression of wheat (Triticum aestivum) TaWRKY1 and TaWRKY33 (a homolog of AtWRKY40) in Arabidopsis enhanced drought tolerance through an ABA-dependent pathway (He et al., 2016). The SlWRKY39 gene, homolog of AtWRKY40, was induced by salt, drought, ABA, SA, JA, and P. syringae (Huang et al., 2012; Sun et al., 2015). The SlWRKY45 gene, another homolog of AtWRKY40, was upregulated by cold treatment (Chen et al., 2015). AtWRKY46 was shown to regulate stress tolerance and hormonal response via ABA signaling and auxin homeostasis (Ding et al., 2015).
Overexpression studies of TaWRKY10 and TaWRKY44 in tobacco showed that these genes acted as enhancers of drought and salt stress tolerance through regulation of osmotic balance and ROS scavenging (Wang et al., 2013, 2015). Overexpression of the Chrysanthemum DgWRKY5 gene enhanced tolerance to salt stress by augmenting ROS scavenging and osmotic adjustment (Liang et al., 2017). The rice OsWRKY30 was involved in drought tolerance in rice via MAPK activation (Rushton et al., 2010; Shen et al., 2012). DgWRKY5, AtWRKY25, TaWRKY44, and OsWRKY30 are all members of the WRKY family Group I (Liang et al., 2017).
The AtWRKY46 gene enhances drought and salt stress tolerance, and regulates stomatal closure (Ding et al., 2015). One of its tomato homologs, SlWRKY41, was upregulated under salt stress, in addition to SlWRKY53, SlWKRY80, and SlWRKY81 (Huang et al., 2012). SlWRKY58 was upregulated under drought stress (Karkute et al., 2018). Overexpression of the cotton (Gossypium hirsutum) GhWRKY41 gene, the closest homolog of SlWRKY58, in tobacco resulted tolerance to drought and salt stress through enhanced stomatal closure as well as by regulating ROS scavenging (Chu et al., 2015).
In addition, altered expression was observed for many other SlWRKY genes in tomato, including induction of SlWRKY23, SlWRKY33, and SlWRKY57 under salt stress (Huang et al., 2012), upregulation of SlWRKY12, SlWRKY13, SlWRKY23, SlWRKY50, and SlWRKY51 under cold stress (Chen et al., 2015), up-regulated SlWRKY31 by drought and salt stress (Huang et al., 2012). Under drought stress, SlWRKY32 and SlWRKY74 were significantly upregulated (Huang et al., 2012), while SlWRKY4 was downregulated (Karkute et al., 2015). The possible positive or negative roles of these SlWKRY genes in plant responses to abiotic stresses still need to be further verified by functional analyses.
WRKYs in Crosstalk Between Abiotic- and Biotic-Stress Tolerance
Several of the aforementioned WRKYs are active at crossroads of plant responses to both biotic and abiotic stresses. In Group I (Figure 1), AtWRKY33 and its two tomato homologs SlWRKY31 and SlWRKY33 are activators of plant defense to several pathogens (Zheng et al., 2006; Lippok et al., 2007; Liu et al., 2014; Li et al., 2015a). In addition, induction of SlWRKY31 and SlWRKY33 was observed under drought and/or salt stresses (Huang et al., 2012). In Group II-a (Figure 2), HvWRKY1 (also designated HvWRKY38 in Mare et al., 2004), AtWRKY40 and its tomato homologs SlWRKY39 and SlWRKY45 are involved in the response to the infection of pathogens and several abiotic stresses (Xu et al., 2006; Shen et al., 2007; Huang et al., 2012; Chen et al., 2015; Sun et al., 2015; Chinnapandi et al., 2017). Similarly, several WRKYs in Group II-b (Figure 2, SlWRKY72 and SlWRKY74) and Group-III (Figure 1, OsWRKY45 and TaWRKY1, SlWRKY80, and SlWRKY81, as well as SlWRKY53 and AtWRKY53) can increase plant tolerance to multiple stresses (Murray et al., 2007; Mukhtar et al., 2008; Qiu and Yu, 2009; Tao et al., 2009, 2011; Bhattarai et al., 2010; Atamian et al., 2012; Huang et al., 2012; Wang et al., 2013, 2015; Marques de Carvalho et al., 2015; He et al., 2016). It is worthwhile to note that WRKYs have been studied for their responses to a single stress at the time. Therefore, further functional analyses of these WRKYs are needed to verify whether the responses to individual stresses remain the same when the plants are exposed to combination(s) of those stress factors. A role for WKRY genes in the interaction of response pathways was obvious in tomato plants in which SlWRKY23 was silenced (Kissoudis, 2016). These plants exhibited increased resistance to tomato powdery caused by Oidium neolycopersici, but this resistance was compromised under salt stress. This example clearly indicates a role for WRKY transcription factors in the crosstalk between biotic and abiotic stress responses, and demonstrates that the responses to individual stresses may not be additive when the plants have to deal with combinatorial stresses.
Tomato is a host for more than 200 species of pathogens, some of which can be controlled by R genes derived from wild tomato relatives (Bai et al., 2018). Evidence is accumulating that plant resistances to pathogens can be attenuated or enhanced by abiotic stresses (Suzuki et al., 2014; Kissoudis et al., 2017). For example, the Mi-1-mediated nematode resistance was compromised under heat stress (Marques de Carvalho et al., 2015). Four tomato WRKYs were shown to contribute to the Mi-1-mediated nematode resistance [SlWRKY72 to SlWRKY74 (Bhattarai et al., 2010) and SlWRKY80 (Atamian et al., 2012)]. The intriguing question is whether these WRKYs are involved in the instability of the Mi-1-mediated resistance under heat stress, or, more generally, do WRKYs play a role in the (in)stability of plant R genes-mediated resistance associated with different molecular mechanisms (Kissoudis et al., 2016).
A (WKRY) gene that confers resistance or tolerance to multiple stresses would be highly useful for breeding. However, WRKY genes can also have opposite effects on abiotic and biotic stress tolerance since complex interactions among signaling networks can lead to both synergistic and antagonistic effects on regulation of plant responses to different stresses (Phukan et al., 2016; Bai et al., 2018). For example, OsWRKY45 that positively mediates broad-spectrum disease resistance while inhibiting adaptation to abiotic stresses (Qiu and Yu, 2009; Tao et al., 2009, 2011), and OsWRKY75 that increases susceptibility to rice blast fungus while improving tolerance to cold stress (Yokotani et al., 2013). Similarly, other transcription factors have also been shown to play an antagonistic role in modulating responses to abiotic and biotic stresses, such as tomato stress-responsive factor TSRF1 (Zhang et al., 2007), Arabidopsis DEAR1 (DREB (dehydration-responsive element binding protein 1) and EAR (ethylene response factor-associated amphiphilic repression) motif protein 1) (Tsutsui et al., 2009). The regulation of plant responses to multiple stresses relies on tightly regulated and highly dynamic regulatory networks where WRKYs can function as activators or repressors (Eulgem and Somssich, 2007; Bakshi and Oelmuller, 2014; Phukan et al., 2016). Therefore, it is necessary that the roles of WRKYs in a plant’s tolerance to biotic and abiotic stresses should be studied under individual stresses as well as combination(s) of the studied stress factors.
It is important to note that some WRKYs were shown to function in a cluster (Cheng et al., 2015; Phukan et al., 2016), such as the AtWRKY18-40-60 cluster (Yan et al., 2013). These three WRKYs form both homomeric and heteromeric complexes to modulate downstream target genes and cross-regulate each other, leading to a variety of responses to stresses and during development. It can be difficult to make use of such WRKY-clusters for crop improvement since multiple responses can lead to unwanted traits along with beneficial effects (Phukan et al., 2016). In tomato, five SlWRKY genes are close homologs of these three AtWRKY genes in Group II-a and shown to be responsive to both abiotic and biotic stresses (Figure 1). Further studies are needed to verify whether they also function in clusters and to identify other SlWRKY clusters. In this review, we tried to infer functions of unstudied SlWRKY genes via their homologs in other plant species. However, it should be stressed that slight changes in the DNA-binding domain may have an important effect on the binding specificity, and sequence homologs may be highly similar yet have different functions (Tao et al., 2009, 2011; Du et al., 2014). For example, the close tomato homologs SlWKRY3 and SlWRKY4 are predicted to interact with the W-box DNA through a different motif, RKYGQK, and WRKYGQK, respectively (Lai et al., 2008; Aamir et al., 2017). There is evidence that motifs outside the WRKY domain may provide binding specificity to WRKYs (Phukan et al., 2016). Also, WRKYs have been shown to bind non-W-box elements, including the sugar-responsive element by HvWRKY46, Calmodulin (CaM)-binding domain and the VQ proteins (Phukan et al., 2016). Identification of motifs associated with functions of tomato WRKYs will contribute to the understanding of their regulatory networks under combined stresses.
Author Contributions
YB designed the outline of the manuscript. YB, SS, and CK contributed to writing and revisions of the manuscript. RV and CvdL contributed to revisions of the manuscript. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00801/full#supplementary-material
FIGURE S1 | The phylogenetic tree of tomato WRKYs and their homologs in Arabidopsis, rice, tobacco, wheat, barley, and grape. WRKYs of tomato (SlWRKYs), Arabidopsis (AtWRKYs), rice (OsWRKYs), tobacco (NtWRKY), wheat (TaWRKY), barley (HvWRKY), cotton (GhWRKY), and grape (VqWKRY) are colored in black, blue, red, green, fuchsia, purple, teal, and olive, respectively. The evolutionary history was inferred by using the Maximum Likelihood method based on the JTT matrix-based model (Jones et al., 1992) and 500 bootstrap (Felsenstein, 1985). The percentages of bootstrap value higher than 50% are indicated on the nodes.
TABLE S1 | WRKY transcription factors discovered in different plant species.
TABLE S2 | The involvements of tomato SlWRKY genes and their homologs in plant responses to biotic and abiotic stresses.
References
Aamir, M., Singh, V. K., Meena, M., Upadhyay, R. S., Gupta, V. K., and Singh, S. (2017). Structural and functional insights into WRKY3 and WRKY4 transcription factors to unravel the WRKY–DNA (W-box) complex interaction in Tomato (Solanum lycopersicum L.). A computational approach. Front. Plant Sci. 8:819. doi: 10.3389/fpls.2017.00819
Adachi, H., Nakano, T., Miyagawa, N., Ishihama, N., Yoshioka, M., Katou, Y., et al. (2015). WRKY transcription factors phosphorylated by MAPK regulate a plant immune NADPH oxidase in Nicotiana benthamiana. Plant Cell 27, 2645–2663. doi: 10.1105/tpc.15.00213
Andreasson, E., Jenkins, T., Brodersen, P., Thorgrimsen, S., Petersen, N. H., Zhu, S., et al. (2005). The MAP kinase substrate MKS1 is a regulator of plant defense responses. EMBO J. 24, 2579–2589. doi: 10.1038/sj.emboj.7600737
Atamian, H. S., Eulgem, T., and Kaloshian, I. (2012). SlWRKY70 is required for Mi-1-mediated resistance to aphids and nematodes in tomato. Planta 235, 299–309. doi: 10.1007/s00425-011-1509-6
Bai, Y., Kissoudis, C., Yan, Z., Visser, R. G. F., and van der Linden, G. (2018). Plant behaviour under combined stress: tomato responses to combined salinity and pathogen stress. Plant J. 93, 781–793. doi: 10.1111/tpj.13800
Bakshi, M., and Oelmuller, R. (2014). WRKY transcription factors: jack of many trades in plants. Plant Signal. Behav. 9:e27700. doi: 10.4161/psb.27700
Bhattarai, K. K., Atamian, H. S., Kaloshian, I., and Eulgem, T. (2010). WRKY72-type transcription factors contribute to basal immunity in tomato and Arabidopsis as well as gene-for-gene resistance mediated by the tomato R gene Mi-1. Plant J. 63, 229–240. doi: 10.1111/j.1365-313X.2010.04232.x
Birkenbihl, R. P., Diezel, C., and Somssich, I. E. (2012). Arabidopsis WRKY33 is a key transcriptional regulator of hormonal and metabolic responses toward Botrytis cinerea infection. Plant Physiol. 159, 266–285. doi: 10.1104/pp.111.192641
Brand, L. H., Fischer, N. M., Harter, K., Kohlbacher, O., and Wanke, D. (2013). Elucidating the evolutionary conserved DNA-binding specificities of WRKY transcription factors by molecular dynamics and in vitro binding assays. Nucleic Acids Res. 41, 9764–9778. doi: 10.1093/nar/gkt732
Chen, L., Yang, Y., Liu, C., Zheng, Y., Xu, M., Wu, N., et al. (2015). Characterization of WRKY transcription factors in Solanum lycopersicum reveals collinearity and their expression patterns under cold treatment. Biochem. Biophys. Res. Commun. 464, 962–968. doi: 10.1016/j.bbrc.2015.07.085
Cheng, H., Liu, H., Deng, Y., Xiao, J., Li, X., and Wang, S. (2015). The WRKY45-2 WRKY13 WRKY42 transcriptional regulatory cascade is required for rice resistance to fungal pathogen. Plant Physiol. 167, 1087–1099. doi: 10.1104/pp.114.256016
Chinnapandi, B., Bucki, P., and Braun Miyara, S. (2017). SlWRKY45, nematode-responsive tomato WRKY gene, enhances susceptibility to the root knot nematode; M. javanica infection. Plant Signal. Behav. 12:e1356530. doi: 10.1080/15592324.2017.1356530
Chu, X., Wang, C., Chen, X., Lu, W., Li, H., Wanr, L., et al. (2015). The cotton WRKY gene GhWRKY41 positively regulates salt and drought stress tolerance in transgenic Nicotiana benthamiana. PLoS One 11:e0157026. doi: 10.1371/journal.pone.0143022
Ding, Z. J., Yan, J. Y., Li, C. X., Li, G. X., Wu, Y. R., and Zheng, S. J. (2015). Transcription factor WRKY46 modulates the development of Arabidopsis lateral roots in osmotic/salt stress conditions via regulation of ABA signaling and auxin homeostasis. Plant J. 84, 56–69. doi: 10.1111/tpj.12958
Dodds, P. N., and Rathjen, J. P. (2010). Plant immunity: towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 11, 539–548. doi: 10.1038/nrg2812
Du, H., Wang, Y., Yang, J., and Yang, W. (2015). Comparative transcriptome analysis of resistant and susceptible tomato lines in response to infection by Xanthomonas perforans race T3. Front. Plant Sci. 6:1173. doi: 10.3389/fpls.2015.01173
Du, M., Zhai, Q., Deng, L., Li, S., Li, H., Yan, L., et al. (2014). Closely related NAC transcription factors of tomato differentially regulate stomatal closure and reopening during pathogen attack. Plant Cell 26, 3167–3184. doi: 10.1105/tpc.114.128272
Eulgem, T., Rushton, P. J., Robatzek, S., and Somssich, I. E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. doi: 10.1016/S1360-1385(00)01600-9
Eulgem, T., and Somssich, I. E. (2007). Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 10, 366–371. doi: 10.1016/j.pbi.2007.04.020
Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x
Franco-Zorrilla, J. M., López-Vidriero, I., Carrasco, J. L., Godoy, M., Vera, P., and Solano, R. (2014). DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc. Natl. Acad. Sci. U.S.A. 111, 2367–2372. doi: 10.1073/pnas.1316278111
Grunewald, W., Karimi, M., Wieczorek, K., Van De Cappelle, E., Wischnitzki, E., Grundler, F., et al. (2008). A role for AtWRKY23 in feeding site establishment of plant-parasitic nematodes. Plant Physiol. 148, 358–368. doi: 10.1104/pp.108.119131
He, G.-H., Xu, J.-Y., Wang, Y.-X., Liu, J.-M., Li, P.-S., Chen, M., et al. (2016). Drought-responsive WRKY transcription factor genes TaWRKY1 and TaWRKY33 from wheat confer drought and/or heat resistance in Arabidopsis. BMC Plant Biol. 16:116. doi: 10.1186/s12870-016-0806-4
Hu, Y., Chen, L., Wang, H., Zhang, L., Wang, F., and Yu, D. (2013). Arabidopsis transcription factor WRKY8 functions antagonistically with its interacting partner VQ9 to modulate salinity stress tolerance. Plant J. 74, 730–745. doi: 10.1111/tpj.12159
Huang, S., Gao, Y., Liu, J., Peng, X., Niu, X., Fei, Z., et al. (2012). Genome-wide analysis of WRKY transcription factors in Solanum lycopersicum. Mol. Genet. Genomics 287, 495–513. doi: 10.1007/s00438-012-0696-6
Huang, Y., Li, M.-Y., Wu, P., Xu, Z.-S., Que, F., Wang, F., et al. (2016). Members of WRKY Group III transcription factors are important in TYLCV defense signaling pathway in tomato (Solanum lycopersicum). BMC Genomics 17:788. doi: 10.1186/s12864-016-3123-2
Ishihama, N., and Yoshioka, H. (2012). Post-translational regulation of WRKY transcription factors in plant immunity. Curr. Opin. Plant Biol. 15, 431–437. doi: 10.1016/j.pbi.2012.02.003
Jones, D. T., Taylor, W. R., and Thornton, J. M. (1992). The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8, 275–282. doi: 10.1093/bioinformatics/8.3.275
Jones, J. D., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Karkute, S. G., Easwaran, M., Gujjar, R. S., Piramanayagam, S., and Singh, M. (2015). Protein modeling and molecular dynamics simulation of SlWRKY4 protein cloned from drought tolerant tomato (Solanum habrochaites) line EC520061. J. Mol. Model. 21:255. doi: 10.1007/s00894-015-2798-7
Karkute, S. G., Gujjar, R. S., Rai, A., Akhtar, M., and Singh, M. (2018). Genome wide expression analysis of WRKY genes in tomato (Solanum lycopersicum) under drought stress. Plant Gene 13, 8–17. doi: 10.1016/j.plgene.2017.11.002
Kim, K.-C., Lai, Z., Fan, B., and Chen, Z. (2008). Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. Plant Cell 20, 2357–2371. doi: 10.1105/tpc.107.055566
Kissoudis, C. (2016). Genetics and Regulation of Combined Abiotic and Biotic Stress Tolerance in Tomato. Ph.D. thesis, Wageningen University, Wageningen.
Kissoudis, C., Seifi, A., Yan, Z., Islam, A. T., van der Schoot, H., van de Wiel, C. C., et al. (2017). Ethylene and abscisic acid signaling pathways differentially influence tomato resistance to combined powdery mildew and salt stress. Front. Plant Sci. 7:2009. doi: 10.3389/fpls.2016.02009
Kissoudis, C., Sunarti, S., van de Wiel, C., Visser, R. G., van der Linden, C. G., and Bai, Y. (2016). Responses to combined abiotic and biotic stress in tomato are governed by stress intensity and resistance mechanism. J. Exp. Bot. 67, 5119–5132. doi: 10.1093/jxb/erw285
Lai, Z., Li, Y., Wang, F., Cheng, Y., Fan, B., Yu, J. Q., et al. (2011). Arabidopsis sigma factor binding proteins are activators of the WRKY33 transcription factor in plant defense. Plant Cell 23, 3824–3841. doi: 10.1105/tpc.111.090571
Lai, Z., Vinod, K., Zheng, Z., Fan, B., and Chen, Z. (2008). Roles of Arabidopsis WRKY3 and WRKY4 transcription factors in plant responses to pathogens. BMC Plant Biol. 8:68. doi: 10.1186/1471-2229-8-68
Li, J., Brader, G., and Palva, E. T. (2004). The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 16, 319–331. doi: 10.1105/tpc.016980
Li, J.-B., Luan, Y.-S., and Liu, Z. (2015a). SpWRKY1 mediates resistance to Phytophthora infestans and tolerance to salt and drought stress by modulating reactive oxygen species homeostasis and expression of defense-related genes in tomato. Plant Cell Tissue Organ Cult. 123, 67–81.
Li, J. B., Luan, Y. S., and Liu, Z. (2015b). Overexpression of SpWRKY1 promotes resistance to Phytophthora nicotianae and tolerance to salt and drought stress in transgenic tobacco. Physiol. Plant. 155, 248–266.
Liang, Q.-Y., Wu, Y.-H., Wang, K., Bai, Z.-Y., Liu, Q.-L., Pan, Y.-Z., et al. (2017). Chrysanthemum WRKY gene DgWRKY5 enhances tolerance to salt stress in transgenic chrysanthemum. Sci. Rep. 7:4799. doi: 10.1038/s41598-017-05170-x
Lippok, B., Birkenbihl, R. P., Rivory, G., Brummer, J., Schmelzer, E., Logemann, E., et al. (2007). Expression of ATWRKY33 encoding a pathogen- or pamp-responsive WRKY transcription factor is regulated by a composite DNA motif containing W-box elements. Mol. Plant Microbe Interact. 20, 420–429. doi: 10.1094/MPMI-20-4-0420
Liu, B., Hong, Y.-B., Zhang, Y.-F., Li, X.-H., Huang, L., Zhang, H.-J., et al. (2014). Tomato WRKY transcriptional factor SlDRW1 is required for disease resistance against Botrytis cinerea and tolerance to oxidative stress. Plant Sci. 227, 145–156. doi: 10.1016/j.plantsci.2014.08.001
Liu, X., Bai, X., Wang, X., and Chu, C. (2007). OsWRKY71, a rice transcription factor, is involved in rice defense response. J. Plant Physiol. 164, 969–979. doi: 10.1016/j.jplph.2006.07.006
Lucioli, A., Perla, C., Berardi, A., Gatti, F., Spanò, L., and Tavazza, M. (2016). Transcriptomics of tomato plants infected with TYLCSV or expressing the central TYLCSV Rep protein domain uncover changes impacting pathogen response and senescence. Plant Physiol. Biochem. 103, 61–70. doi: 10.1016/j.plaphy.2016.02.034
Mao, G., Meng, X., Liu, Y., Zheng, Z., Chen, Z., and Zhang, S. (2011). Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 23, 1639–1653. doi: 10.1105/tpc.111.084996
Marques de Carvalho, L., Benda, N. D., Vaughan, M. M., Cabrera, A. R., Hung, K., Cox, T., et al. (2015) Mi-1-mediated nematode resistance in tomatoes is broken by short-term heat stress but recovers over time. J. Nematol. 47, 133–140.
Mare, C., Mazzucotelli, E., Crosatti, C., Francia, E., and Cattivelli, L. (2004). Hv-WRKY38: a new transcription factor involved in cold-and drought-response in barley. Plant Mol. Biol. 55, 399–416. doi: 10.1007/s11103-004-0906-7
Mukhtar, M. S., Deslandes, L., Auriac, M. C., Marco, Y., and Somssich, I. E. (2008). The Arabidopsis transcription factor WRKY27 influences wilt disease symptom development caused by Ralstonia solanacearum. Plant J. 56, 935–947. doi: 10.1111/j.1365-313X.2008.03651.x
Murray, S. L., Ingle, R. A., Petersen, L. N., and Denby, K. J. (2007). Basal resistance against Pseudomonas syringae in Arabidopsis involves WRKY53 and a protein with homology to a nematode resistance protein. Mol. Plant Microbe Interact. 20, 1431–1438. doi: 10.1094/MPMI-20-11-1431
Pandey, S. P., and Somssich, I. E. (2009). The role of WRKY transcription factors in plant immunity. Plant Physiol. 150, 1648–1655. doi: 10.1104/pp.109.138990
Peng, Y., Bartley, L. E., Chen, X., Dardick, C., Chern, M., Ruan, R., et al. (2008). OsWRKY62 is a negative regulator of basal and Xa21-mediated defense against Xanthomonas oryzae pv. oryzae in rice. Mol. Plant 1, 446–458. doi: 10.1093/mp/ssn024
Phukan, U. J., Jeena, G. S., and Shukla, R. K. (2016). WRKY transcription factors: molecular regulation and stress responses in plants. Front. Plant Sci. 7:760. doi: 10.3389/fpls.2016.00760
Qiu, J. L., Fiil, B. K., Petersen, K., Nielsen, H. B., Botanga, C. J., Thorgrimsen, S., et al. (2008). Arabidopsis MAP kinase 4 regulates gene expression through transcription factor release in the nucleus. EMBO J. 27, 2214–2221. doi: 10.1038/emboj.2008.147
Qiu, Y., and Yu, D. (2009). Over-expression of the stress-induced OsWRKY45 enhances disease resistance and drought tolerance in Arabidopsis. Environ. Exp. Bot. 65, 35–47. doi: 10.1016/j.envexpbot.2008.07.002
Rezzonico, F., Rupp, O., and Fahrentrapp, J. (2017). Pathogen recognition in compatible plant-microbe interactions. Sci. Rep. 7:6383. doi: 10.1038/s41598-017-04792-5
Rinerson, C. I., Rabara, R. C., Tripathi, P., Shen, Q. J., and Rushton, P. J. (2015). The evolution of WRKY transcription factors. BMC Plant Biol. 15:66. doi: 10.1186/s12870-015-0456-y
Rushton, P. J., Somssich, I. E., Ringler, P., and Shen, Q. J. (2010). WRKY transcription factors. Trends Plant Sci. 15, 247–258. doi: 10.1016/j.tplants.2010.02.006
Sarris, P. F., Duxbury, Z., Huh, S. U., Ma, Y., Segonzac, C., Sklenar, J., et al. (2015). A plant immune receptor detects pathogen effectors that target WRKY transcription factors. Cell 161, 1089–1100. doi: 10.1016/j.cell.2015.04.024
Shen, H., Liu, C., Zhang, Y., Meng, X., Zhou, X., Chu, C., et al. (2012). OsWRKY30 is activated by MAP kinases to confer drought tolerance in rice. Plant Mol. Biol. 80, 241–253. doi: 10.1007/s11103-012-9941-y
Shen, Q.-H., Saijo, Y., Mauch, S., Biskup, C., Bieri, S., Keller, B., et al. (2007). Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 315, 1098–1103. doi: 10.1126/science.1136372
Sun, X.-C., Gao, Y.-F., Li, H.-R., Yang, S.-Z., and Liu, Y.-S. (2015). Over-expression of SlWRKY39 leads to enhanced resistance to multiple stress factors in tomato. J. Plant Biol. 58, 52–60. doi: 10.1007/s12374-014-0407-4
Suzuki, N., Rivero, R.-M., Shulaev, V., Blumwald, E., and Mittler, R. (2014). Abiotic and biotic stress combinations. New Phytol. 203, 32–43. doi: 10.1111/nph.12797
Tao, Z., Kou, Y., Liu, H., Li, X., Xiao, J., and Wang, S. (2011). OsWRKY45 alleles play different roles in abscisic acid signalling and salt stress tolerance but similar roles in drought and cold tolerance in rice. J. Exp. Bot. 62, 4863–4874. doi: 10.1093/jxb/err144
Tao, Z., Liu, H., Qiu, D., Zhou, Y., Li, X., Xu, C., et al. (2009). A pair of allelic WRKY genes play opposite roles in rice-bacteria interactions. Plant Physiol. 151, 936–948. doi: 10.1104/pp.109.145623
Tsutsui, T., Kato, W., Asada, Y., Sako, K., Sato, T., Sonoda, Y., et al. (2009). DEAR1, a transcriptional repressor of DREB protein that mediates plant defense and freezing stress responses in Arabidopsis. J. Plant Res. 122, 633–643. doi: 10.1007/s10265-009-0252-6
Van Esse, H. P., Fradin, E. F., De Groot, P. J., De Wit, P. J., and Thomma, B. P. (2009). Tomato transcriptional responses to a foliar and a vascular fungal pathogen are distinct. Mol. Plant Microbe Interact. 22, 245–258. doi: 10.1094/MPMI-22-3-0245
Wang, C., Deng, P., Chen, L., Wang, X., Ma, H., Hu, W., et al. (2013). A wheat WRKY transcription factor TaWRKY10 confers tolerance to multiple abiotic stresses in transgenic tobacco. PLoS One 8:e65120. doi: 10.1371/journal.pone.0065120
Wang, X., Guo, R., Tu, M., Wang, D., Guo, C., Wan, R., et al. (2017). Ectopic expression of the wild grape WRKY transcription factor VqWRKY52 in Arabidopsis thaliana enhances resistance to the biotrophic pathogen powdery mildew but not to the necrotrophic pathogen Botrytis cinerea. Front. Plant Sci. 8:97. doi: 10.3389/fpls.2017.00097
Wang, X., Zeng, J., Li, Y., Rong, X., Sun, J., Sun, T., et al. (2015). Expression of a WRKY44, a wheat WRKY gene, in transgenic tobacco confers multiple abiotic stress tolerances. Front. Plant Sci. 6:615. doi: 10.3389/fpls.2015.00615
Xing, D.-H., Lai, Z.-B., Zheng, Z.-Y., Vinod, K., Fan, B.-F., and Chen, Z.-X. (2008). Stress-and pathogen-induced Arabidopsis WRKY48 is a transcriptional activator that represses plant basal defense. Mol. Plant 1, 459–470. doi: 10.1093/mp/ssn020
Xu, X., Chen, C., Fan, B., and Chen, Z. (2006). Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell 18, 1310–1326. doi: 10.1105/tpc.105.037523
Yan, L., Liu, Z.-Q., Xu, Y.-H., Lu, K., Wang, X.-F., and Zhang, D.-P. (2013). Auto- and cross-repression of three Arabidopsis WRKY transcription factors WRKY18, WRKY40, and WRKY60 negatively involved in ABA signaling. J. Plant Growth Regul. 32, 399–416. doi: 10.1007/s00344-012-9310-8
Yang, Y., Chi, Y., Wang, Z., Zhou, Y., Fan, B., and Chen, Z. (2016). Functional analysis of structurally related soybean GmWRKY58 and GmWRKY76 in plant growth and development. J. Exp. Bot. 67, 4727–4742. doi: 10.1093/jxb/erw252
Yokotani, N., Sato, Y., Tanabe, S., Chujo, T., Shimizu, T., Okada, K., et al. (2013). WRKY76 is a rice transcriptional repressor playing opposite roles in blast disease resistance and cold stress tolerance. J. Exp. Bot. 64, 5085–5097. doi: 10.1093/jxb/ert298
Zhang, H., Li, W., Chen, J., Yang, Y., Zhang, Z., Zhang, H., et al. (2007). Transcriptional activator TSRF1 reversely regulates pathogen resistance and osmotic stress tolerance in tobacco. Plant Mol. Biol. 63, 63–71. doi: 10.1007/s11103-006-9072-4
Zhang, Y., and Wang, L. (2005). The WRKY transcription factor superfamily: its origin in eukaryotes and expansion in plants. BMC Evol. Biol. 5:1.
Zheng, Z., Qamar, S. A., Chen, Z., and Mengiste, T. (2006). Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 48, 592–605. doi: 10.1111/j.1365-313X.2006.02901.x
Keywords: abiotic stress, biotic stress, combined stresses, disease resistance, effector-triggered immunity (ETI), PAMP-triggered immunity (PTI)
Citation: Bai Y, Sunarti S, Kissoudis C, Visser RGF and van der Linden CG (2018) The Role of Tomato WRKY Genes in Plant Responses to Combined Abiotic and Biotic Stresses. Front. Plant Sci. 9:801. doi: 10.3389/fpls.2018.00801
Received: 16 January 2018; Accepted: 24 May 2018;
Published: 13 June 2018.
Edited by:
Jean-benoit Morel, Institut National de la Recherche Agronomique Centre Montpellier, FranceReviewed by:
Sang-Soo Kwak, Korea Research Institute of Bioscience and Biotechnology (KRIBB), South KoreaRakesh Kumar Shukla, Central Institute of Medicinal and Aromatic Plants (CIMAP), India
Copyright © 2018 Bai, Sunarti, Kissoudis, Visser and van der Linden. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuling Bai, bai.yuling@wur.nl
 Yuling Bai
Yuling Bai Sri Sunarti
Sri Sunarti Christos Kissoudis
Christos Kissoudis Richard G. F. Visser
Richard G. F. Visser C. G. van der Linden
C. G. van der Linden