- 1Laboratory of Biochemistry, Wageningen University and Research, Wageningen, Netherlands
- 2Microbial Physiology, Groningen Biomolecular Sciences and Biotechnology Institute, University of Groningen, Groningen, Netherlands
- 3Department of Biology and Biotechnology, University of Pavia, Pavia, Italy
- 4Biomolecular Mass Spectrometry and Proteomics, Bijvoet Center for Biomolecular Research and Utrecht Institute for Pharmaceutical Research, Utrecht University, Utrecht, Netherlands
3-Hydroxybenzoate 6-hydroxylase (3HB6H, EC 1.13.14.26) is a FAD-dependent monooxygenase involved in the catabolism of aromatic compounds in soil microorganisms. 3HB6H is unique among flavoprotein hydroxylases in that it harbors a phospholipid ligand. The purified protein obtained from expressing the gene encoding 3HB6H from Rhodococcus jostii RHA1 in the host Escherichia coli contains a mixture of phosphatidylglycerol and phosphatidylethanolamine, which are the major constituents of E. coli’s cytoplasmic membrane. Here, we purified 3HB6H (RjHB6H) produced in the host R. jostii RHA#2 by employing a newly developed actinomycete expression system. Biochemical and biophysical analysis revealed that Rj3HB6H possesses similar catalytic and structural features as 3HB6H, but now contains phosphatidylinositol, which is a specific constituent of actinomycete membranes. Native mass spectrometry suggests that the lipid cofactor stabilizes monomer-monomer contact. Lipid analysis of 3HB6H from Pseudomonas alcaligenes NCIMB 9867 (Pa3HB6H) produced in E. coli supports the conclusion that 3HB6H enzymes have an intrinsic ability to bind phospholipids with different specificity, reflecting the membrane composition of their bacterial host.
Introduction
Rhodococcus jostii RHA1 is a biotechnologically and environmentally important bacterium from the order Actinomycetales. Together with the genera Nocardia, Corynebacterium and Mycobacterium, Rhodococcus forms a distinct group of bacteria called mycolata (Finnerty, 1992; Brennan and Nikaido, 1995; Chun et al., 1996; Gürtler et al., 2004), characterized by a complex cell envelope (Sutcliffe, 1998; Guerin et al., 2010; De Carvalho et al., 2014) and an impressive catabolic diversity, allowing adaptation to different carbon sources for growth (van der Geize and Dijkhuizen, 2004). In comparison with other mycolata, R. jostii RHA1 is particularly rich in oxygenases (203 putative genes) and ligases (192 putative genes), gained primarily through ancient gene duplications or acquisitions (McLeod et al., 2006; Yam et al., 2010).
We recently reported the crystal structure of R. jostii RHA1 3-hydroxybenzoate 6-hydroxylase (3HB6H), produced as a recombinant protein in Escherichia coli (Montersino et al., 2013). 3HB6H (EC 1.13.14.26) is a NADH and FAD-dependent monooxygenase that catalyzes the para-hydroxylation of 3-hydroxybenzoate to 2,5-dihydroxybenzoate, using a Tyr-His pair for substrate binding and catalysis (Sucharitakul et al., 2015). The crystal structure analysis revealed that 3HB6H has the conserved fold of group A flavoprotein hydroxylases (Montersino et al., 2011; Huijbers et al., 2014), but differs from the other family members in additional binding of phospholipids. The tightly bound phospholipids were identified as a mixture of PG and PE, which are the major constituents of the E. coli cytoplasmic membrane (Pulfer and Murphy, 2003; Oursel et al., 2007). The fatty acyl chains of the phospholipid ligands of 3HB6H protrude into the substrate-binding pockets, whereas the surface-exposed hydrophilic headgroups are involved in enzyme dimerization (Figure 1) (Montersino et al., 2013).
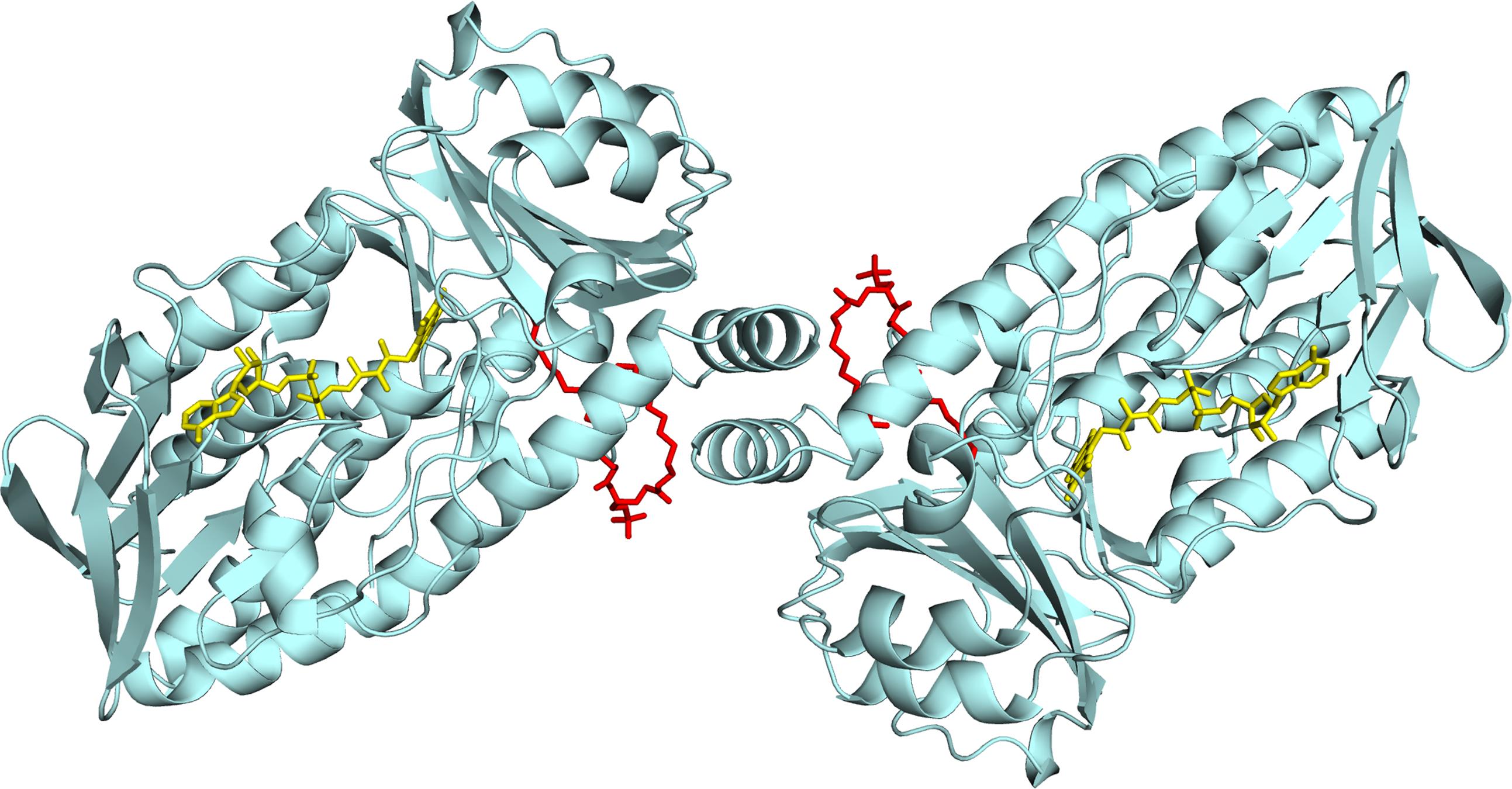
FIGURE 1. Lipid binding in 3HB6H from R. jostii RHA1. Cartoon of the three-dimensional structure of the 3HB6H dimer (Montersino et al., 2013). The protein is shown in light blue and the FAD cofactor is depicted as stick model in yellow. The lipid cofactor and the aromatic substrate are shown as stick models in red and dark blue, respectively.
To shed more light on the role of these lipid guests, bearing in mind the different lipid compositions of Gram-positive and Gram-negative bacterial membranes (Finnerty, 1992; Sutcliffe, 1998), in the present work we produced Rj3HB6H in a newly developed R. jostii RHA1#2 expression strain and, in addition, 3HB6H from Pseudomonas alcaligenes NCIMB 9867 (Pa3HB6H) in E. coli. Biochemical and biophysical characterization revealed that Rj3HB6H possesses similar catalytic and structural features as 3HB6H, but contains PI as glycerophospholipid ligand. Lipid analysis of Pa3HB6H indicates that lipid binding is an intrinsic property of prokaryotic 3-hydroxybenzoate 6-hydroxylases.
Materials and Methods
Chemicals
Aromatic compounds were purchased from Sigma-Aldrich (St Louis, MO, United States) and Acros Organics (Morris Plains, NJ, United States). Catalase, FAD, FMN, arabinose, antibiotics, Terrific broth (TB) and LB broth (Miller) (LB) were from Sigma-Aldrich (St Louis, MO, United States). Pefabloc SC and DNase I were obtained from Roche Diagnostics GmbH (Mannheim, Germany). Restriction enzymes and Pfu DNA polymerase were from Thermo Fischer Scientific (United States). 4-androstene-3,17-dione was from Merck (Oss, Netherlands). Crystallization kits were purchased from New Hampton (Aliso Viejo, CA, United States). Immobilized metal affinity chromatography columns (His GraviTrap) were from GE Healthcare Bioscience AB (Uppsala, Sweden). All other chemicals were from commercial sources and of the purest grade available.
Bacterial Strains and Primers
All bacterial strains and primers used in this study are listed in Tables 1, 2.
Construction of Rhodococcus Expression Vector Q2+
The E. coli-Rhodococcus shuttle vector pRESQ (van der Geize et al., 2002) was modified by insertion of the RP4 oriT of pK18mobsacB (Schäfer et al., 1994) enabling trans-conjugal transfer of the resulting vector. For this, the oriT-region was amplified from pK18mobsacB by PCR using forward primer oriT-F and reverse primer oriT-R (Table 1). The obtained 549 bp PCR-product was cloned into the SmaI-site of pRESQ, resulting in pQmob. A duplicate region of 424 bp on pQmob was removed by deleting the 760 bp XbaI-BspHI fragment, yielding pQmobΔd. The egfp gene from pIJ8630 (Sun et al., 1999) was amplified by PCR using forward primer egfp-PciI-F, containing a PciI restriction site, and reverse primer egfp-PciI-R, also containing a PciI restriction site (Table 1). The 744 bp PciI-PciI fragment containing the egfp gene was cloned into the PciI-site of pQmobΔd to generate peGFPQ.
The R. jostii strain RHA1 genomic region consisting of gene ro00440, its promoter region and the prmA promoter (PprmA) (here referred to as region reg1-PprmA; GenBank accession number CP000431: nt 521345 - nt 523358) was amplified from genomic DNA of R. jostii RHA1 by PCR using forward primer reg1-BglII-F, containing a BglII restriction site, and reverse primer prmA-NdeI-R, containing an NdeI restriction site (Table 1). The 2014 bp BglII-NdeI reg1-PprmA fragment was cloned into the BglII-NdeI sites of peGFPQ, yielding prMOeGFPQ1.
The R. jostii RHA1 gene ro00452 and its promoter region (here referred to as region reg2; CP000431: nt 534363 – nt 536227) were amplified by PCR using forward primer reg2-PciI-F, containing a PciI restriction site and reverse primer reg2-PciI-R, also containing a PciI restriction site (Table 1). The 1880 bp PciI-PciI reg2 fragment was cloned into the PciI-site of prMOeGFPQ1, resulting in prMOeGFPQ2.
For construction of expression vector Q2+, the egfp gene of prMOeGFPQ2 was replaced with a multiple cloning site (MCS). For this, part of the MCS of pBluescript KS was amplified by PCR using forward primer MCS-NdeI-F, containing an NdeI restriction site and reverse primer MCS-PciI-R, containing a PciI restriction site (Table 1). The 125 bp NdeI-PciI MCS fragment was cloned into the NdeI–PciI site of prMOeGFPQ2, replacing the NdeI–PciI region containing egfp, resulting in the expression vector Q2+.
Cloning and Production of 3HB6H in R. jostii RHA1#2
The 1321 bp NdeI-XmnI fragment of pBAD-3HB6H-His6 (Montersino and van Berkel, 2012) containing the 3HB6H gene including the His6-tag, was cloned into the NdeI-HindIII (Klenow-fragment treated) site of expression vector Q2+ to generate Q2+-3HB6H-His6.
Rhodococcus cells were electroporated as described previously (van der Geize et al., 2000). Prior to electroporation, plasmid DNA was desalted by dialyzing 10 μL plasmid DNA for 30 min on a Millipore “V” Series filter disk (0.025 μm) floating on MiliQ water.
Cultures of R. jostii RHA1#2 were grown in LB broth supplemented with 50 μg⋅mL-1 kanamycin at 30°C at 200 rpm. R. jostii RHA1#2 cells harboring Q2+3HB6H-His6 were grown overnight in 3 mL LB broth, diluted 1:300 in 300 mL LB broth in a 3 L Erlenmeyer flask and grown for 20–24 h. Cultures were induced by adding 2 mM 4-androstene-3,17-dione dissolved in acetone. Growth was continued for 48 h after induction. Cells were harvested by centrifugation at 4°C and pellets were washed once with ice-cold 20 mM potassium phosphate, pH 7.2, containing 300 mM NaCl. After centrifugation at 4°C, cells were stored at -20°C.
Cloning and Production of 3HB6H from Pseudomonas alcaligenes NCIMB 9867
The xlnD gene sequence encoding for Pa3HB6H (UniProt: Q9F131) was synthesized and subcloned in a pBAD vector by GeneArt (Invitrogen, Carlsbad, CA, United States). The resulting construct (pBAD-Pa3HB6H-His6) was verified by automated sequencing of both strands and electroporated into E. coli TOP10 cells for recombinant expression.
For enzyme production, E. coli TOP10 cells, harboring the pBAD-Pa3HB6H-His6 plasmid, were grown in TB medium at 37°C supplemented with 100 μg⋅mL-1 ampicillin until an optical density (OD600 nm) of 0.8 was reached. Expression was induced by the addition of 0.02% (w/v) arabinose and incubation was continued for 40 h at 17°C. Cells were harvested by centrifugation at 4°C and stored at -20°C.
Enzyme Purification
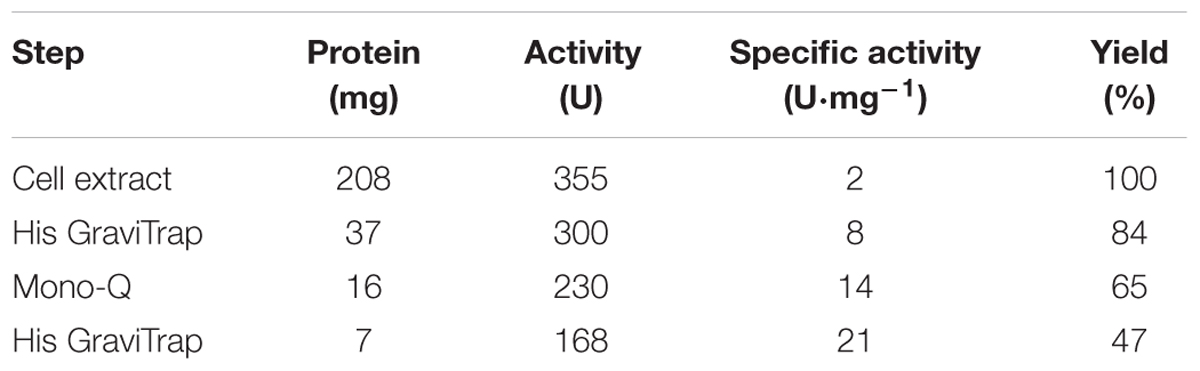
Rj3HB6H was purified to apparent homogeneity using an Åkta Explorer chromatography system (GE-Healthcare). R. jostii RHA1#2 cells containing the recombinant protein were suspended in ice-cold 20 mM potassium phosphate, pH 7.2, containing 300 mM NaCl, 1 mM Pefabloc SC, 1 mg DNAse and 100 μM MgCl2, and subsequently passed twice through a precooled French Pressure cell (SLM Aminco, SLM Instruments, Urbana, IL, United States) at 16,000 psi. The resulting homogenate was centrifuged at 25,000 × g for 45 min at 4°C to remove cell debris, and the supernatant was applied onto a Ni-NTA agarose column (16 mm × 50 mm) equilibrated with 20 mM potassium phosphate, pH 7.2, containing 300 mM NaCl. After washing with five volumes of equilibration buffer, the enzyme was eluted with 300 mM imidazole in equilibration buffer. The resulting Rj3HB6H fraction was supplemented with 100 μM FAD and loaded onto a Source Q-15 anion exchange column (16 mm × 90 mm), pre-equilibrated with 50 mM Bis-Tris, 0.1 mM EDTA, pH 7.2. After washing with two volumes of equilibrating buffer, the enzyme was eluted with a linear gradient of 0–1 M NaCl in the same buffer. Active fractions were pooled, concentrated to 10 mg⋅mL-1 using ultrafiltration (Amicon 30 kDa cutoff filter), and applied onto a Superdex S-200 (26 mm × 600 mm) column running in 50 mM potassium phosphate, 150 mM NaCl, pH 7.2. Active fractions were concentrated to 10 mg⋅mL-1 using ultrafiltration (Amicon 30 kDa cutoff filter) and dialyzed at 4°C against 50 mM Bis-Tris, pH 7.2. The final Rj3HB6H preparation showed a single band after SDS-PAGE. The specific activity of the purified enzyme was 21 U mg-1 using the standard activity assay (Table 3A).
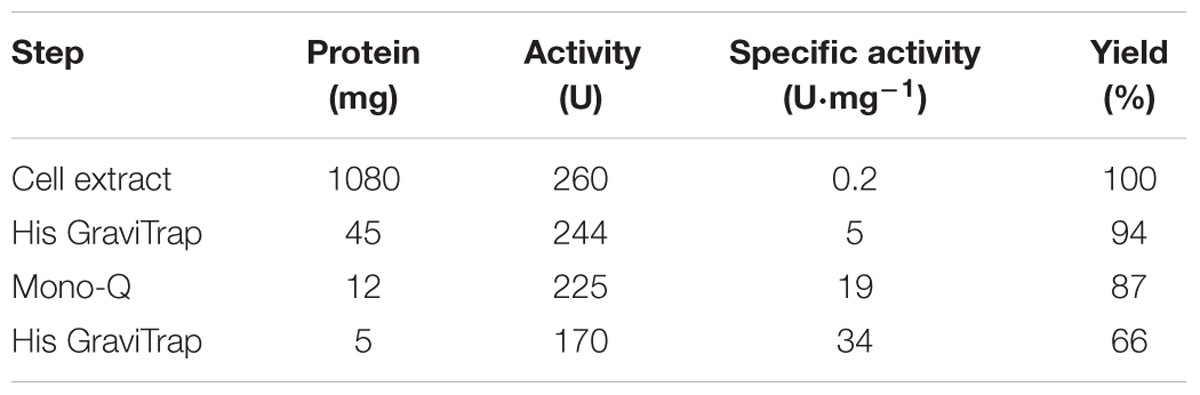
Pa3HB6H was purified to apparent homogeneity, applying essentially the same procedure as described above for Rj3HB6H. The final Pa3HB6H preparation showed a single band after SDS-PAGE. The specific activity of the purified enzyme was 34 U⋅mg-1 using the standard activity assay (Table 3B).
Purified enzymes were flash frozen in 1 mL aliquots in liquid nitrogen and stored at -80°C. Before use, thawed enzyme samples were incubated with 0.1 mM FAD and excess FAD was removed using a gel filtration column (10 mm × 100 mm) containing Bio-Gel P-6DG.
Biochemical Characterization
Molar absorption coefficients of protein-bound FAD were determined from absorption spectra of Rj3HB6H and Pa3HB6H recorded in the presence and absence of 0.1% (w/v) SDS, assuming a molar absorption coefficient for free FAD of 11.3 mM-1⋅cm-1 at 450 nm. The enzyme concentration of Rj3HB6H was determined by measuring the absorbance at 453 nm using a molar absorption coefficient for protein-bound FAD of 10.3 mM-1⋅cm-1. The enzyme concentration of Pa3HB6H was determined by measuring the absorbance at 450 nm using a molar absorption coefficient for protein-bound FAD of 11.0 mM-1⋅cm-1. Rj3HB6H and Pa3HB6H activity was determined at 25°C by measuring NADH consumption at 360 nm (Montersino and van Berkel, 2012). The standard assay mixture contained 50 mM Tris-SO4, pH 8.0, 200 μM 4-hydroxybenzoate and 250 μM NADH. Steady-state kinetic parameters were determined from measurements at 25°C in 50 mM Tris-SO4, pH 8.0. Hydroxylation efficiencies were determined by oxygen consumption experiments, essentially as described before (Montersino and van Berkel, 2012).
Crystallization and Structure Determination
Crystals of Rj3HB6H for structure determination were obtained by the sitting drop vapor diffusion method at 20°C by mixing equal volumes (2 μL) of protein and reservoir solutions. Protein solutions consisted of 30 mg⋅mL-1 enzyme in 1 mM FAD, 2 mM 3-hydroxybenzoate, and 50 mM Bis-Tris, pH 7.2, whereas precipitant solutions consisted of 30% PEG 4000, 0.2 M lithium sulfate, and 0.1 M Tris-HCl, pH 8.5. Yellow crystals grew in 1 day.
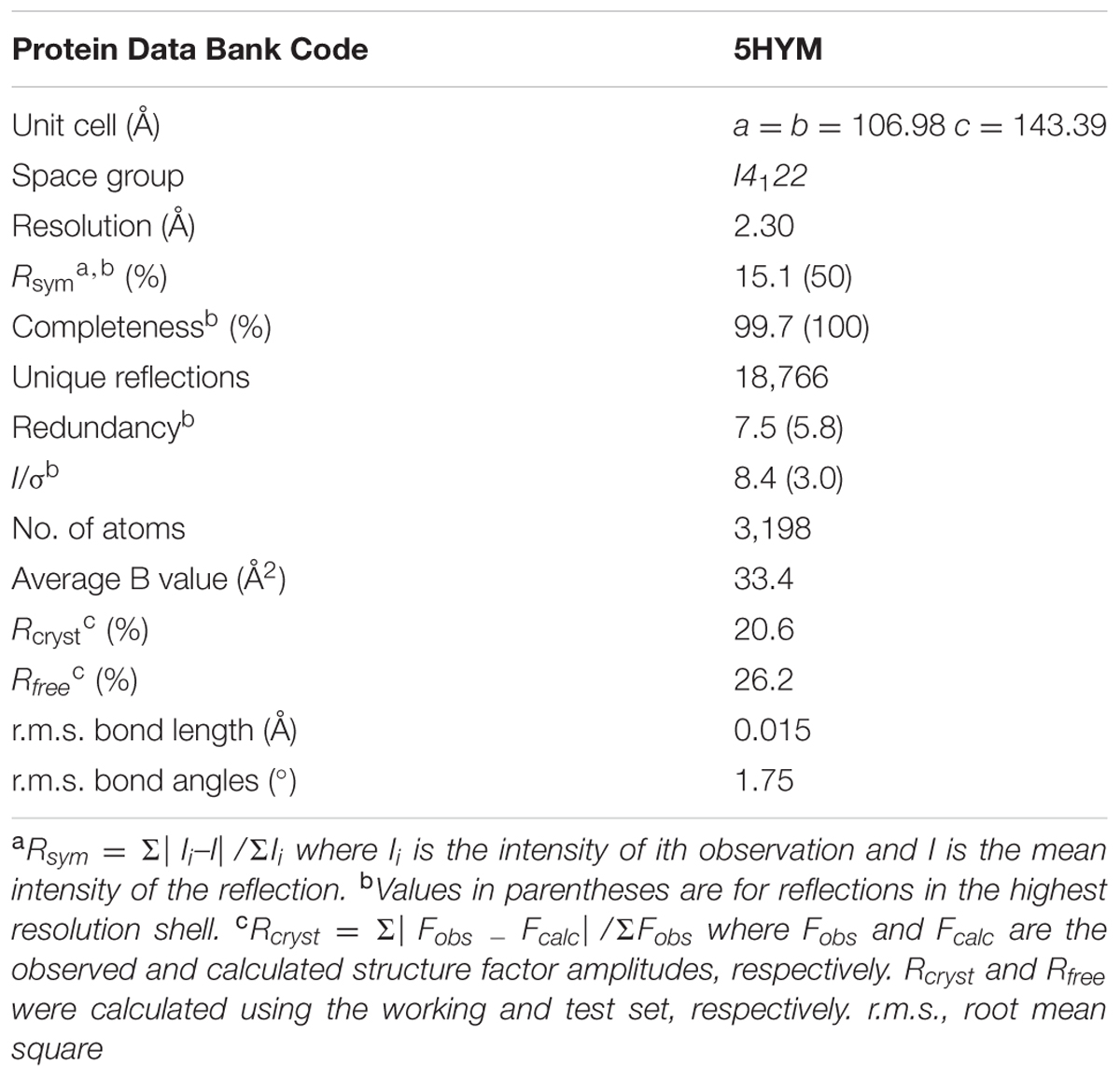
X-ray diffraction data were collected at Grenoble and processed with the CCP4 package (Winn et al., 2011). The Rj3HB6H structure was solved by molecular replacement using the structure of a monomer of 3HB6H (pdb entry: 4BJZ) as search model. Crystallographic computing and model analysis were performed with COOT (Emsley et al., 2010), PHENIX (Adams et al., 2010) and the CCP4 package (Potterton et al., 2004). Pictures were generated with Pymol (Schrodinger, 2015) and CCP4 (Potterton et al., 2004). Data collection parameters and refinement statistics are presented in Table 4.
The atomic coordinates and structure factors of Rj3HB6H (code 5HYM) have been deposited in the Protein Data Bank1.
Lipid Identification and Native ESI-MS Experiments
Extraction and identification of protein-bound lipids from Rj3HB6H and Pa3HB6H was performed as described for 3HB6H (Montersino et al., 2013). For nanoflow ESI-MS analysis under native conditions, enzyme samples were prepared in 50 mM ammonium acetate, pH 6.8. For analysis under denaturing conditions, enzyme samples were diluted either in 50% acetonitrile with 0.2% formic acid or in 5% formic acid. Native MS analysis was performed using a LC-T nanoflow ESI orthogonal TOF mass spectrometer (Micromass, Manchester, United Kingdom) in positive ion mode with a capillary voltage of 1.3 kV. The cone voltage was varied between 90 and 150 V and source pressure was set to 6.9 mbar to enhance transmission of large ions. Lipid identification was performed using a Quattro Ultima nanoflow triple quadrupole mass spectrometer (Micromass, Manchester, United Kingdom) in negative ion mode, with a capillary voltage of 1.3 kV and a cone voltage of 150 V. For MS/MS analysis, argon was supplied in the collision cell (2.0 × 10-3 bar). Collision energy was adjusted to gain optimal fragmentation. Both mass spectrometers were equipped with a Z-spray nano-electrospray ionization source. Measurements were performed by using gold-coated needles, made from borosilicate glass capillaries (Kwik-Fill; World precision Instruments, Sarasota) on a P-97 puller (from Sutter Instruments, Novato, CA, United States). Needles were coated with a gold layer using an Edwards Scancoat six Pirani 501 sputter coater (Edwards laboratories, Milpitas, CA, United States). All TOF spectra were mass calibrated by using an aqueous solution of cesium iodide (25 mg⋅mL-1).
Sequence Comparison
Protein sequences were retrieved using protein resources from the National Centre for Biotechnology Information2 and UniProt Database3. Multiple sequence alignments were made using CLUSTALW (Thompson et al., 1994). Phylogenetic plots were made using FigTree4.
Results
Biochemical Properties of Rj3HB6H
Expression of the 3HB6H gene from R. jostii RHA1#2 yielded about 7 mg of purified Rj3HB6H protein from 10 g wet cells (Table 3A). Rj3HB6H displayed the same absorption spectrum as 3HB6H, with maxima at 274, 383, and 453 nm and a shoulder at 480 nm (Montersino and van Berkel, 2012). A molar absorption coefficient of protein-bound flavin, 𝜀453 = 10.3 mM-1 cm-1, was used for both proteins.
Determination of steady-state kinetic parameters revealed that Rj3HB6H behaves similarly as 3HB6H using 3-hydroxybenzoate as variable substrate and fixed NADH concentration (kcat = (20 ± 1) s-1; KM = (35 ± 3) μM; kcat/KM = (5.7 ± 0.8) × 105 s-1⋅M-1) and with variable concentration of NADH (preferred coenzyme) and fixed 3-hydroxybenzoate concentration (kcat = (20 ± 1) s-1; KM = (68 ± 5) μM; kcat/KM = (3.0 ± 0.4) × 105 s-1⋅M-1). Rj3HB6H displays a very low NADH oxidase activity (<1 U⋅mg-1). Uncoupling of hydroxylation of 3-hydroxybenzoate occurs to a minor extent (less than 10%), while 2,5-dihydroxybenzoate is a strong non-substrate effector (kcat = (6 ± 0.8) s-1; KM = (150 ± 30) μM; kcat/KM = (4.0 ± 1.3) × 104 s-1⋅M-1), efficiently stimulating the rate of flavin reduction by NADH (Montersino and van Berkel, 2012; Sucharitakul et al., 2012, 2013; Ni et al., 2016).
Structural Characterization
Rj3HB6H crystals grew in similar conditions as found for 3HB6H, and are isomorphous to those of 3HB6H, where lithium sulfate was present instead of sodium acetate. The three-dimensional structure of Rj3HB6H was solved at 2.3 Å resolution by molecular replacement (Table 4). The isoalloxazine moiety of FAD was refined with full occupancy in the in conformation. Similar to the crystallographic analysis of 3HB6H, no substrate could be detected in the active site of the enzyme, despite presence of excess 3-hydroxybenzoate in the crystallization drop. The protein crystallizes as a dimer, just as 3HB6H (Montersino et al., 2013), and contains a phospholipid molecule in each subunit. The electron density of the phospholipid in the crystal structure was refined as two acyl chains, one of twelve and one of seventeen carbon units. Superimposition of the Rj3HB6H and 3HB6H models (root mean square deviation = 0.22 Å) shows minor deviations (Figure 2A). The phospholipid is located in a tunnel, which runs from the dimer interface to the active site (Figure 2B), and interacts with the opposite monomer. The phosphate group resides at the protein surface near Arg350 and Lys385, and the electron density of the headgroup is consistent with the presence of a cyclohexanehexol moiety (Figure 3A).
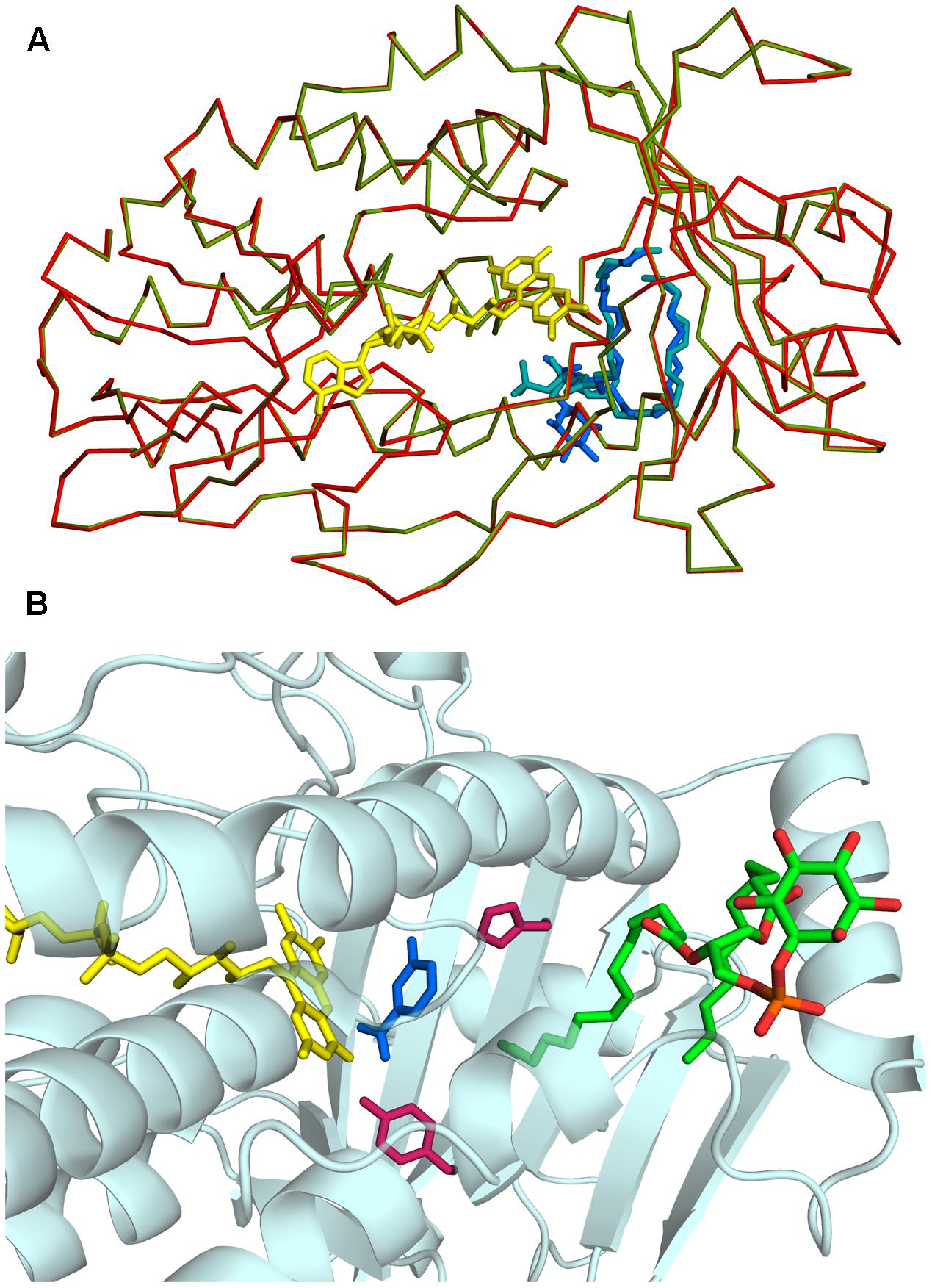
FIGURE 2. Three-dimensional structure of Rj3HB6H. (A) View of α-carbon traces of the refined structures of 3HB6H (green; pdb code 4BJZ) and Rj3HB6H (red; pdb code 5HYM), following superposition of corresponding main chain atoms. FAD is shown as a stick model in yellow. The lipid ligands are shown as stick models in shades of blue in the back of the protein. (B) The phospholipid is located in a tunnel, which runs from the dimer interface to the active site. The FAD cofactor is depicted in yellow. The lipid cofactor is colored by elements, and the active site residues His213 and Tyr217 are in dark red. The 3-hydroxybenzoate in blue is a superposition from the structure of the 3HB6H variant H213S, which contains bound substrate (pdb code 4bk1).
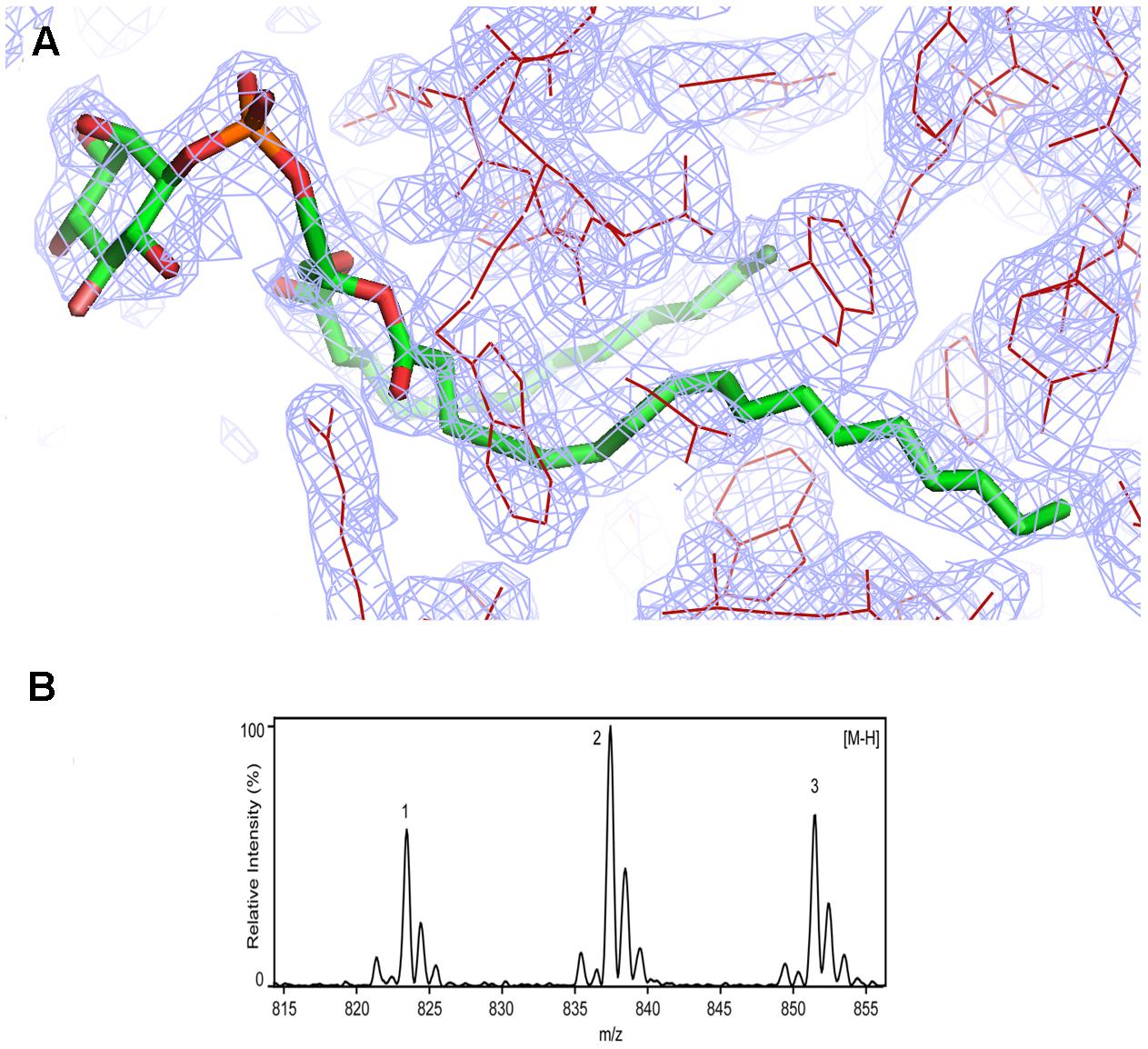
FIGURE 3. Identification of phosphatidylinositol in Rj3HB6H. (A) Weighted (2Fo-Fc) electron-density map of the lipid cofactor. (B) ESI-MS spectrum of lipid extract collected in negative mode. For peak assignment, see “Results” section.
Identification of Protein-Bound Lipid Molecules
Assignment of protein-bound phospholipids was achieved by ESI-MS analysis of the low molecular weight components extracted from denatured Rj3HB6H. The mass spectrum in negative mode (Figure 3B) displayed three main peaks with m/z values of 823, 837, and 851. From the MS pattern it was evident that Rj3HB6H binds a phospholipid with a bigger headgroup compared to that of the lipid found in 3HB6H.
Fragmentation analysis and comparison of data to reference lipid MS spectra led to a match of the obtained mass peaks with those of PI, having aliphatic chains containing 15 to 19 carbons. Peak 1 (m/z 823) is assigned to PI (15:0/18:0), peak 2 (m/z 837) is assigned to PI (16:0/18:0) (Sharp et al., 2007; Morita et al., 2011) and peak 3 ((m/z 851) is assigned to PI (16:0/19:0) with alternate acylate form (tuberculostearic acid) (Drage et al., 2010).
Typical fragmentation of PI was visible in the MS/MS spectra by signature peaks with m/z values of 153, 223, 241, and 297, representing glycerol phosphate water (m/z 153) and inositol headgroup fragments (Lang and Philp, 1998; Pulfer and Murphy, 2003; Oursel et al., 2007) (data not shown). Minor peaks at approximately 2 m/z values lower than the identified peaks represent the same PI, containing one unsaturated bond.
Protein Oligomeric Composition
To gain further insight into the enzyme–lipid interaction, we determined the oligomeric protein composition of 3HB6H and Rj3HB6H using native MS (Leney and Heck, 2017). As a first step, we determined the experimental masses of the denatured proteins. The measured values (46,766 ± 4 Da for 3HB6H and 46,761 ± 2 Da for Rj3HB6H) agree with the mass deduced from the primary sequence, lacking the N-terminal methionine.
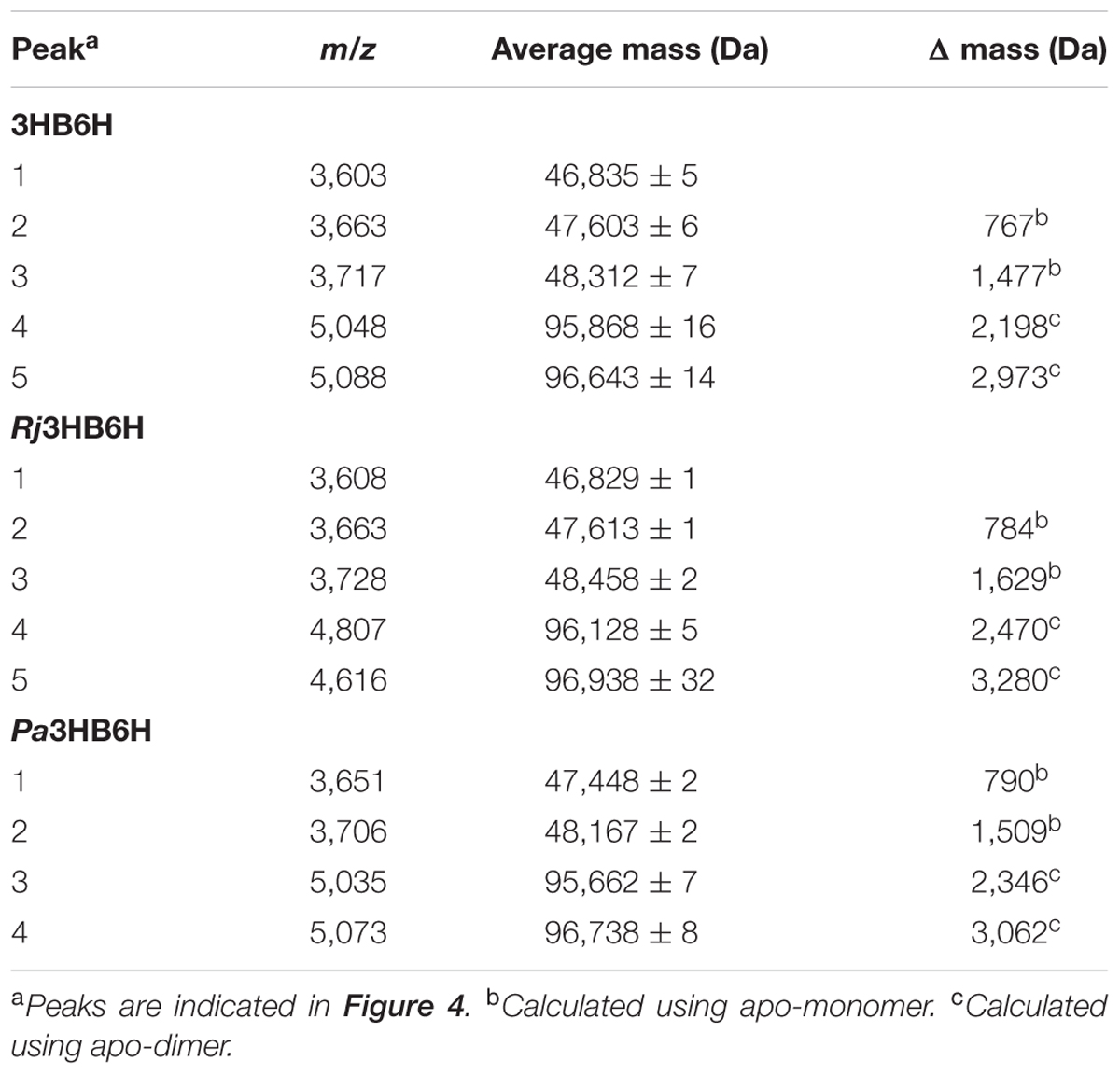
Native MS of 3HB6H showed eight charge states corresponding to five different protein forms (Figure 4A and Table 5). The charge state distribution ions +12, +13, and +14 represent the monomeric apoprotein (average mass 46,835 ± 5 Da; red stars), the monomeric holoprotein (average mass 47603 ± 6 Da; green stars), and the monomeric holoprotein containing additionally one PG/PE molecule (average mass 48,312 ± 7 Da; blue stars). The charge state distribution ions +18, +19, +20, +21, and +22 predominantly represent the dimeric holoprotein with either one or two PG/PE molecules bound (average mass 95,868 ± 16 Da; orange stars, and 96,643 ± 14 Da; purple stars, respectively). Tandem MS experiments revealed that one, two, three, or four ligands can be expelled from 3HB6H. The assignment of bound ligands was made on the basis of total mass increase and comparison with the mass of the native apoprotein (Table 5).
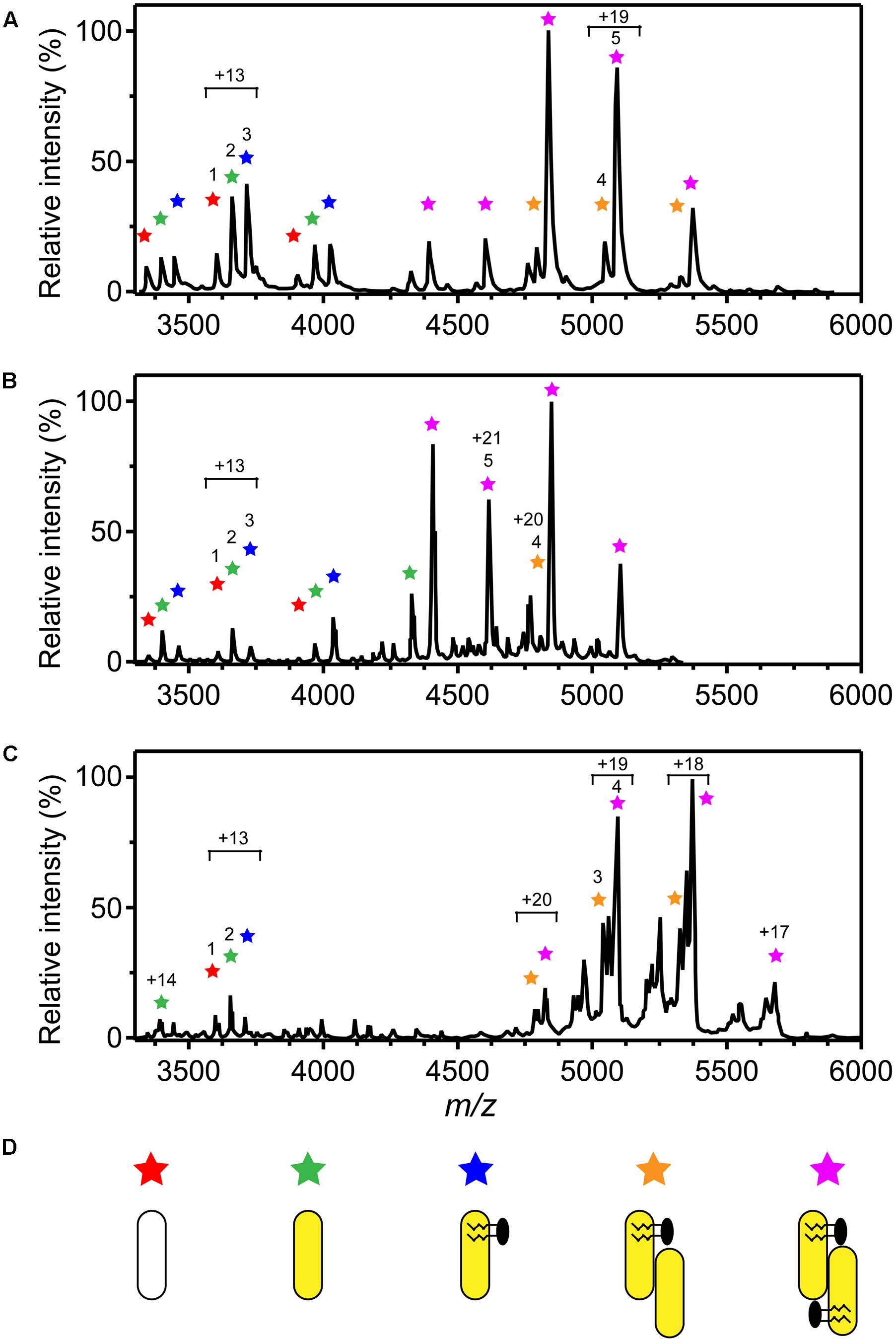
FIGURE 4. Oligomer distribution and lipid composition of 3HB6H enzymes as determined by native ESI-MS. Mass spectra were recorded in 50 mM ammonium acetate, pH 6.8. (A) Mass spectrum of 3HB6H. (B) Mass spectrum of Rj3HB6H. (C) Mass spectrum of Pa3HB6H. Masses and intensities of numbered peaks are listed in Table 5. (D) Cartoons of the various subunit compositions. Apoprotein is indicated in white, holoprotein in yellow and lipid molecules in black.
Native MS of Rj3HB6H also showed a range of charge state distributions (Figure 4B and Table 5). The charge state distribution ions +12, +13, and +14 represent the monomeric apoprotein (average mass 46,829 ± 1 Da; red star), the monomeric holoprotein (average mass 47613 ± 1 Da; green stars), and the monomeric holoprotein containing one PI molecule (average mass 48,458 ± 2 Da; blue star). The latter species differs from the related 3HB6H species (Figure 4A, blue stars) because it has a bigger lipid headgroup. The major species in the native mass spectrum of Rj3HB6H corresponds to the holo-Rj3HB6H dimer with PI bound to both subunits (average mass 96,938 ± 32 Da; Figure 4B, yellow stars). Only by magnification it is possible to detect a minor peak representing the holo-Rj3HB6H dimer with one PI bound (average mass 96,128 ± 5 Da; Figure 4B, orange stars). A cartoon of the different subunit compositions of 3HB6H is presented in Figure 4D.
Conservation of Lipid Binding Site
To analyze whether the lipid-binding site of 3HB6H is conserved among species, we explored the natural diversity of 3HB6H enzymes. 3HB6H activity has been reported for Gram-positive and Gram-negative bacteria and for yeasts. Besides from the R. jostii prototype, the enzymes from Klebsiella pneumonia M5a1 (Suárez et al., 1995; Liu et al., 2005), Pseudomonas alcaligenes NCIMB 9867 (Gao et al., 2005), Polaromonas naphthalenivorans CJ2 (Park et al., 2007), Corynebacterium glutamicum ATCC 12032 (Yang et al., 2010), Rhodococcus sp. NCIMB 12038 (Liu et al., 2011) and Candida parapsilosis (Holesova et al., 2011) have been characterized to some extent.
From the structural data of the R. jostii 3HB6H enzyme and the multiple sequence alignment presented in Figure 5 it can be inferred that residues directly involved in lipid binding in Rj3HB6H are not always conserved in the orthologs; among the bacterial enzymes studied, most sequence divergence occurs in 3HB6H from P. alcaligenes NCIMB 9867 (Pa3HB6H). This prompted us to study the lipid binding properties of the Pseudomonas 3HB6H enzyme.
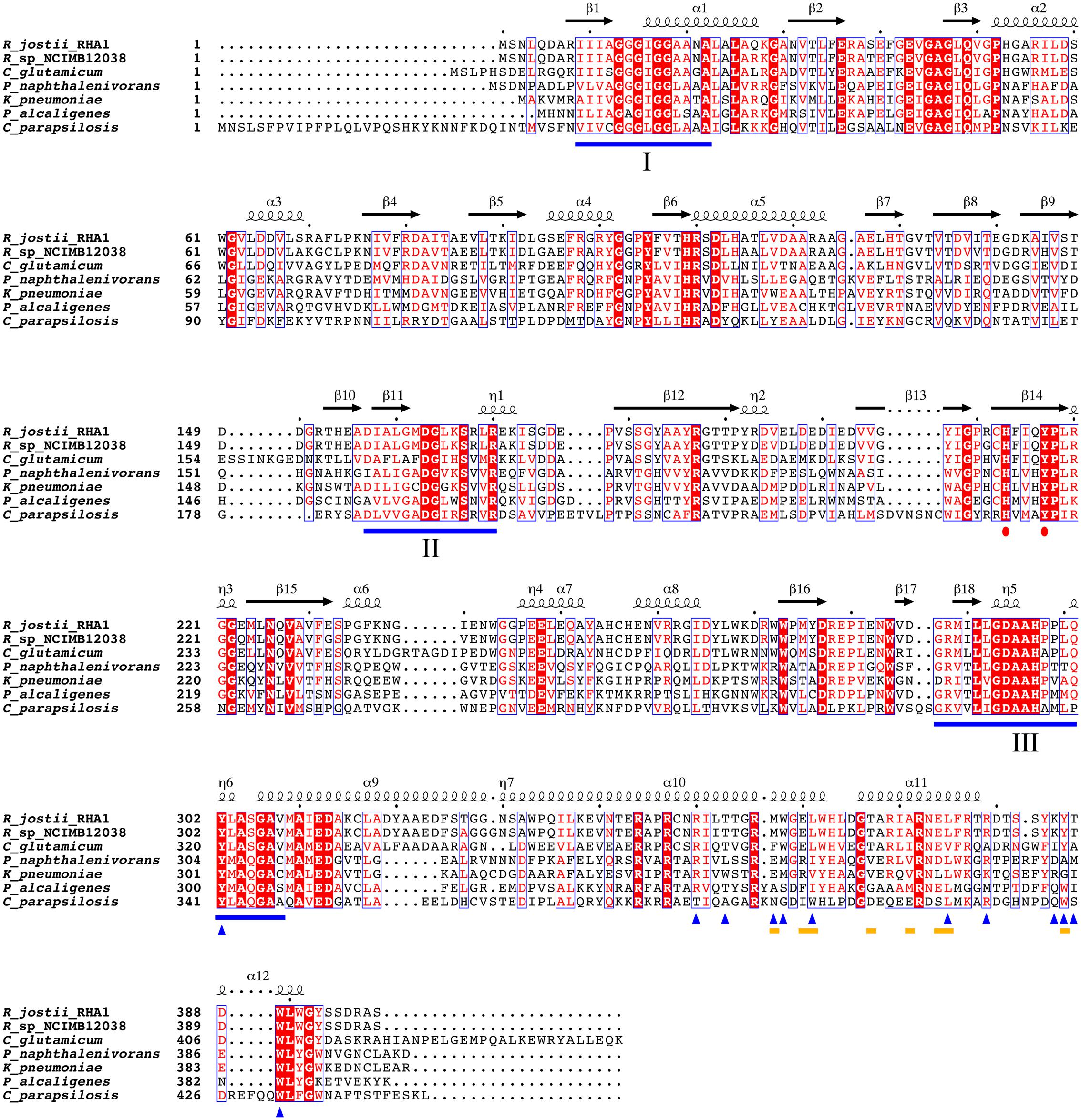
FIGURE 5. Multiple sequence alignment of known 3HB6H enzymes. UniProt ID numbers: Q0SFK6, R. jostii RHA1; E7CYP8, Rhodococcus sp. NCIMB 12038; Q8NLB6, C. glutamicum ATCC 12032; Q3S4B7, P. naphthalenivorans CJ2; Q6EXK1, K. pneumonia M5a1; Q9F131, P. alcaligenes NCIMB 9867; CPAG_03410, C. parapsilosis. Identical residues are shown in red. Flavin binding motifs are underlined in blue [I: GXGXXG; II: DG; III: GD (Eppink et al., 1997)]. The His-Tyr pair involved in substrate binding and hydroxylation is marked with red dots. The yellow lines mark residues involved in dimerization contacts. Blue triangles indicate residues involved in lipid binding. Secondary structure assigned from the 3HB6H crystal structure (4BK1). Diagram was produced using ESPript (Robert and Gouet, 2014).
Expression of the Pa3HB6H gene in E. coli TOP10 cells yielded about 10 mg of enzyme from a 1 L batch culture. Purified Pa3HB6H had a specific activity of 34 U⋅mg-1 (Table 3B) and migrated in SDS-PAGE as a single band with an apparent subunit mass of 47 kDa (not shown). ESI-MS established that native Pa3HB6H is a dimer, and not a trimer as suggested earlier (Gao et al., 2005), and that the enzyme indeed contains lipids (Figure 4C and Table 5). The mass spectrum of extracted lipids showed peaks with m/z values characteristic of PG and PE with aliphatic chains ranging from 14 to 19 carbons, similar to the previously identified lipids in 3HB6H from R. jostii RHA1 produced in E. coli (Montersino et al., 2013).
Discussion
3HB6H is a flavoenzyme that catalyzes the para-hydroxylation of 3-hydroxybenzoate to gentisate, a key step in the catabolism of lignin-derived aromatic compounds in the soil (Pérez-Pantoja et al., 2010). Up to now, 3HB6H is the only flavoprotein monooxygenase that has been found to bind phospholipids (Montersino et al., 2013). Structural analysis showed that the hydrophobic tails of the phospholipids deeply penetrate into the substrate-binding domains, whereas the hydrophilic parts are exposed on the protein surface, connecting the dimerization domains (Figure 1). Attempts to obtain native lipid-free protein were not successful, indicating that the phospholipids are important to attain a properly folded protein (Montersino et al., 2013).
3HB6H binds a mixture of PG and PE, the major constituents of the E. coli inner membrane (Montersino et al., 2013). By expressing its gene in R. jostii RHA1#2, we aimed at unraveling the lipid binding abilities of 3HB6H in the original host. Although E. coli gives considerable higher yields (Montersino and van Berkel, 2012), significant quantities of soluble His-tagged Rj3HB6H were obtained. The difference in enzyme yield could be linked to the type of induction and promoter strength used in the R. jostii RHA1#2 strain, which is based on the propane monooxygenase operon (Sharp et al., 2007). Nevertheless, our results show that the newly developed R. jostii RHA1#2 strain opens new prospects for actinomycetes as host cells for production of recombinant proteins (Nakashima et al., 2005).
Rj3HB6H displayed similar catalytic and structural properties as 3HB6H, and the mode of lipid binding was highly conserved (Figure 2). Gratifyingly, the crystallographic data and mass spectrometry analysis provided clear evidence that Rj3HB6H contains PI as natural glycerophospholipid cofactor (Figure 3). The crystal structure showed that the inositol headgroups of the phospholipids are located at the protein surface, and that the sn-2 acyl moieties are in contact with helix 11 of the other subunit (Figure 1). Based on MS/MS analysis, we identified the bound phospholipids as a mixture of PIs with carbon chains between 15 and 19 carbons. One of the extracted lipids was identified as tuberculostearic acid, an alternative acylated form of palmitate present in the membranes of Rhodococcus and Mycobacterium (Drage et al., 2010).
Rj3HB6H is a dimer both in solution and in crystal form, but native MS showed a ratio of monomer to dimer of about 1:3 (Figure 4B). Release of only one PI from the dimer resulted in monomerization in the gas phase. A similar observation was made with 3HB6H, but with this enzyme more dimers containing only one bound lipid (PG or PE) were detected (Figure 4A). 3HB6H dimers containing two phospholipids seem to be more stable in the gas phase than dimers containing one phospholipid. This strongly supports that lipid binding near the dimer interface stabilizes monomer contacts.
Native MS-analysis showed that Pa3HB6H is a homodimer and not a trimer as postulated earlier (Gao et al., 2005). The dimeric nature is in agreement with the structural properties of 3HB6H and Rj3HB6H. MS-analysis also revealed that recombinant Pa3HB6H binds the same type of phospholipids as 3HB6H. This supports that lipid binding is an intrinsic property of 3HB6Hs. As a main result, it appears that the 3HB6H family uses phospholipids as a common tool to increase their dimerization strength. Phospholipid binding is independent of the type of lipid headgroup, but relies on the presence of hydrophobic tunnels running from the protein surface to the active site.
Like PG/PE in E. coli (Oursel et al., 2007), PI is the major lipid membrane component in Rhodococcus (Nigou et al., 2003). This may explain why PG/PE are found as lipid ligands of 3HB6H and Pa3HB6H, while PI is found in Rj3HB6H. PI is the precursor for lipoarabinomannan and PI-mannoside synthesis. Glycolipid synthesis and reorganization of membrane composition allow Rhodococcus to adapt to environmental changes (Lang and Philp, 1998; Sharp et al., 2007; Guerin et al., 2010; Morita et al., 2011; De Carvalho et al., 2014). Binding of PI may localize 3HB6H at the cytoplasmic membrane, via inositol recognition of other proteins or specific phospholipid patching on the inner side of the membrane (Morita et al., 2011). At those specific spots, uptake of aromatic compounds from the environment may be coupled more efficiently to their catabolism.
Taking together, phospholipids do not have a direct catalytic role in 3HB6H, but are involved in stabilizing the dimer contact and, possibly, substrate orientation (Montersino et al., 2013). At this stage, we cannot exclude that bound phospholipids have some other function, for instance in directing the cytoplasmic membrane localization or in guiding/protecting molecules from entering the active site. In addition, the R. jostii RHA1#2 expression strain described here represents a useful alternative for the production of (whole-cell) biocatalysts.
Author Contributions
WvB and LD initiated the project; SM, EtP, AW, AB, AH, RvG, LD, AM, and WvB designed experiments and analyzed data; EtP constructed Rhodococcus expression vector Q2+; RO crystallized Rj3HB6H and determined the crystal structure; SM, AW, and AB performed analytical and biochemical experiments; SM, EtP, AM, AW, and WvB wrote the manuscript.
Funding
This study was supported by the Integrated Biosynthesis Organic Synthesis program (IBOS; project number 053.63.013) of the Netherlands Organization for Scientific Research (NWO). Additional support from Proteins At Work (project 184.032.201), a program of the Netherlands Proteomics Centre financed by the Netherlands Organization for Scientific Research (NWO) as part of the National Roadmap Large-scale Research Facilities of the Netherlands is acknowledged.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
Part of the content of this manuscript has been published in the Ph.D. thesis of Dr. Stefania Montersino (Montersino, 2012).
Footnotes
- ^ http://pdbe.org/5HYM
- ^ www.ncbi.nlm.nih.gov
- ^ http://www.uniprot.orgwww.uniprot.org
- ^ tree.bio.ed.ac.uk
Abbreviations
3HB6H, recombinant 3-hydroxybenzoate 6-hydroxylase from Rhodococcus jostii RHA1 produced in Escherichia coli; Pa3HB6H, recombinant 3-hydroxybenzoate 6-hydroxylase from Pseudomonas alcaligenes NCIMB 9867 produced in Escherichia coli; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PI, phosphatidylinositol; Rj3HB6H, recombinant 3-hydroxybenzoate 6-hydroxylase from Rhodococcus jostii RHA1 produced in Rhodococcus jostii RHA1#2.
References
Adams, P. D., Afonine, P. V., Bunkóczi, G., Chen, V. B., Davis, I. W., Echols, N., et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. 66(Pt 2), 213–221. doi: 10.1107/s0907444909052925
Brennan, P. J., and Nikaido, H. (1995). The envelope of mycobacteria. Annu. Rev. Biochem. 64, 29–63. doi: 10.1146/Annurev.Bi.64.070195.000333
Chun, J., Kang, S. O., Hah, Y. C., and Goodfellow, M. (1996). Phylogeny of mycolic acid-containing actinomycetes. J. Ind. Microbiol. 17, 205–213. doi: 10.1007/Bf01574694
De Carvalho, C. C., Costa, S. S., Fernandes, P., Couto, I., and Viveiros, M. (2014). Membrane transport systems and the biodegradation potential and pathogenicity of genus Rhodococcus. Front. Physiol. 5:133. doi: 10.3389/fphys.2014.00133
Drage, M. G., Tsai, H. C., Pecora, N. D., Cheng, T. Y., Arida, A. R., Shukla, S., et al. (2010). Mycobacterium tuberculosis lipoprotein LprG (Rv1411c) binds triacylated glycolipid agonists of Toll-like receptor 2. Nat. Struct. Mol. Biol. 17, 1088–1095. doi: 10.1038/nsmb.1869
Emsley, P., Lohkamp, B., Scott, W. G., and Cowtan, K. (2010). Features and development of Coot. Acta Crystallogr. D Biol. 66, 486–501. doi: 10.1107/S0907444910007493
Eppink, M., Schreuder, H., and van Berkel, W. (1997). Identification of a novel conserved sequence motif in flavoprotein hydroxylases with a putative dual function in FAD/NAD(P)H binding. Protein Sci. 6, 2454–2458. doi: 10.1002/pro.5560061119
Finnerty, W. R. (1992). The biology and genetics of the genus Rhodococcus. Annu. Rev. Microbiol. 46, 193–218. doi: 10.1146/Annurev.Micro.46.1.193
Gao, X. L., Tan, C. L., Yeo, C. C., and Poh, C. L. (2005). Molecular and biochemical characterization of the xlnD-encoded 3-hydroxybenzoate6-hydroxylase involved in the degradation of 2,5-xylenol via the gentisate pathway in Pseudomonas alcaligenes NCIMB 9867. J. Bacteriol. 187, 7696–7702. doi: 10.1128/JB.187.22.7696-7702.2005
Guerin, M. E., Korduláková, J., Alzari, P. M., Brennan, P. J., and Jackson, M. (2010). Molecular basis of phosphatidyl-myo-inositol mannoside biosynthesis and regulation in mycobacteria. J. Biol. Chem. 285, 33577–33583. doi: 10.1074/jbc.R110.168328
Gürtler, V., Mayall, B. C., and Seviour, R. (2004). Can whole genome analysis refine the taxonomy of the genus Rhodococcus? FEMS Microbiol. Rev. 28, 377–403. doi: 10.1016/j.femsre.2004.01.001
Holesova, Z., Jakubkova, M., Zavadiakova, I., Zeman, I., Tomaska, L., and Nosek, J. (2011). Gentisate and 3-oxoadipate pathways in the yeast Candida parapsilosis: identification and functional analysis of the genes coding for 3-hydroxybenzoate 6-hydroxylase and 4-hydroxybenzoate 1-hydroxylase. Microbiology 157, 2152–2163. doi: 10.1099/mic.0.048215-0
Huijbers, M. M. E., Montersino, S., Westphal, A. H., Tischler, D., and van Berkel, W. J. H. (2014). Flavin dependent monooxygenases. Arch. Biochem. Biophys. 544, 2–17. doi: 10.1016/j.abb.2013.12.005
Lang, S., and Philp, J. C. (1998). Surface-active lipids in rhodococci. Antonie van Leeuwenhoek 74, 59–70. doi: 10.1023/A:1001799711799
Leney, A. C., and Heck, A. J. R. (2017). Native mass spectrometry: what is in the name? J. Am. Soc. Mass Spectrom. 28, 5–13. doi: 10.1007/s13361-016-1545-3
Liu, D. Q., Liu, H., Gao, X. L., Leak, D. J., and Zhou, N. Y. (2005). Arg(169) is essential for catalytic activity of 3-hydroxybenzoate 6-hydroxylase from Klebsiella pneumoniae M5a1. Microbiol. Res. 160, 53–59. doi: 10.1016/j.micres.2004.09.003
Liu, T. T., Xu, Y., Liu, H., Luo, S., Yin, Y. J., Liu, S. J., et al. (2011). Functional characterization of a gene cluster involved in gentisate catabolism in Rhodococcus sp. strain NCIMB 12038. Appl. Microbiol. Biotechnol. 90, 671–678. doi: 10.1007/s00253-010-3033-1
McLeod, M. P., Warren, R. L., Hsiao, W. W. L., Araki, N., Myhre, M., Fernandes, C., et al. (2006). The complete genome of Rhodococcus sp RHA1 provides insights into a catabolic powerhouse. Proc. Natl. Acad. Sci. U.S.A. 103, 15582–15587. doi: 10.1073/pnas.0607048103
Montersino, S. (2012). Structural and Biochemical Characterization of 3-Hydroxybenzoate 6-Hydroxylase. Doctoral thesis, Wageningen University, Wageningen.
Montersino, S., Orru, R., Barendregt, A., Westphal, A. H., van Duijn, E., Mattevi, A., et al. (2013). Crystal structure of 3-hydroxybenzoate 6-hydroxylase uncovers lipid-assisted flavoprotein strategy for regioselective aromatic hydroxylation. J. Biol. Chem. 288, 26235–26245. doi: 10.1074/jbc.M113.479303
Montersino, S., Tischler, D., Gassner, G. T., and van Berkel, W. J. H. (2011). Catalytic and structural features of flavoprotein hydroxylases and epoxidases. Adv. Synth. Catal. 353, 2301–2319. doi: 10.1002/adsc.201100384
Montersino, S., and van Berkel, W. J. H. (2012). Functional annotation and characterization of 3-hydroxybenzoate 6-hydroxylase from Rhodococcus jostii RHAl. Biochim. Biophys. Acta 1824, 433–442. doi: 10.1016/j.bbapap.2011.12.003
Morita, Y. S., Fukuda, T., Sena, C. B. C., Yamaryo-Botte, Y., McConville, M. J., and Kinoshita, T. (2011). Inositol lipid metabolism in mycobacteria: biosynthesis and regulatory mechanisms. Biochim. Biophys. Acta 1810, 630–641. doi: 10.1016/j.bbagen.2011.03.017
Nakashima, N., Mitani, Y., and Tamura, T. (2005). Actinomycetes as host cells for production of recombinant proteins. Microb. Cell Fact. 4:7. doi: 10.1186/1475-2859-4-7
Ni, Y., Fernandez-Fueyo, E., Baraibar, A. G., Ullrich, R., Hofrichter, M., Yanase, H., et al. (2016). Peroxygenase-catalyzed oxyfunctionalization reactions promoted by the complete oxidation of methanol. Angew. Chem. Int. Ed. Engl. 55, 798–801. doi: 10.1002/anie.201507881
Nigou, J., Gilleron, M., and Puzo, G. (2003). Lipoarabinomannans: from structure to biosynthesis. Biochimie 85, 153–166. doi: 10.1016/S0300-9084(03)00048-8
Oursel, D., Loutelier-Bourhis, C., Orange, N., Chevalier, S., Norris, V., and Lange, C. M. (2007). Lipid composition of membranes of Escherichia coli by liquid chromatography/tandem mass spectrometry using negative electrospray ionization. Rapid Commun. Mass Spectrom. 21, 1721–1728. doi: 10.1002/rcm.3013
Park, M., Jeon, Y., Jang, H. H., Ro, H.-S., Park, W., Madsen, E. L., et al. (2007). Molecular and biochemical characterization of 3-hydroxybenzoate 6-hydroxylase from Polaromonas naphthalenivorans CJ2. Appl. Environ. Microbiol. 73, 5146–5152. doi: 10.1128/AEM.00782-07
Pérez-Pantoja, D., González, B., and Pieper, D. H. (2010). “Aerobic Degradation of aromatic hydrocarbons,” in Handbook of Hydrocarbon and Lipid Microbiology, ed. K. N. Timmis (Berlin: Springer), 799–837. doi: 10.1007/978-3-540-77587-4_60
Potterton, L., McNicholas, S., Krissinel, E., Gruber, J., Cowtan, K., Emsley, P., et al. (2004). Developments in the CCP4 molecular-graphics project. Acta Crystallogr. D Biol. 60, 2288–2294. doi: 10.1107/S0907444904023716
Pulfer, M., and Murphy, R. C. (2003). Electrospray mass spectrometry of phospholipids. Mass Spectrom. Rev. 22, 332–364. doi: 10.1002/mas.10061
Robert, X., and Gouet, P. (2014). Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324. doi: 10.1093/nar/gku316
Schäfer, A., Tauch, A., Jäger, W., Kalinowski, J., Thierbach, G., and Pühler, A. (1994). Small mobilizable multipurpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145, 69–73. doi: 10.1016/0378-1119(94)90324-7
Sharp, J. O., Sales, C. M., LeBlanc, J. C., Liu, J., Wood, T. K., Eltis, L. D., et al. (2007). An inducible propane monooxygenase is responsible for N-nitrosodimethylamine degradation by Rhodococcus sp strain RHA1. Appl. Environ. Microbiol. 73, 6930–6938. doi: 10.1128/AEM.01697-07
Suárez, M., Ferrer, E., Garridopertierra, A., and Martín, M. (1995). Purification and characterization of the 3-hydroxybenzoate 6-hydroxylase from Klebsiella pneumoniae. FEMS Microbiol. Lett. 126, 283–290. doi: 10.1111/J.1574-6968.1995.Tb07431.X
Sucharitakul, J., Medhanavyn, D., Pakotiprapha, D., van Berkel, W. J., and Chaiyen, P. (2015). Tyr217 and His213 are important for substrate binding and hydroxylation of 3-hydroxybenzoate 6-hydroxylase from Rhodococcus jostii RHA1. FEBS J. 283, 860–881. doi: 10.1111/febs.13636
Sucharitakul, J., Tongsook, C., Pakotiprapha, D., van Berkel, W. J. H., and Chaiyen, P. (2013). The reaction kinetics of 3-hydroxybenzoate 6-hydroxylase from Rhodococcus jostii RHA1 provide an understanding of the para-hydroxylation enzyme catalytic cycle. J. Biol. Chem. 288, 35210–35221. doi: 10.1074/jbc.M113.515205
Sucharitakul, J., Wongnate, T., Montersino, S., van Berkel, W. J. H., and Chaiyen, P. (2012). Reduction kinetics of 3-hydroxybenzoate 6-hydroxylase from Rhodococcus jostii RHA1. Biochemistry 51, 4309–4321. doi: 10.1021/bi201823c
Sun, J. H., Kelemen, G. H., Fernández-Abalos, J. M., and Bibb, M. J. (1999). Green fluorescent protein as a reporter for spatial and temporal gene expression in Streptomyces coelicolor A3(2). Microbiology 145, 2221–2227. doi: 10.1099/00221287-145-9-2221
Sutcliffe, I. C. (1998). Cell envelope composition and organisation in the genus Rhodococcus. Antonie van Leeuwenhoek 74, 49–58. doi: 10.1023/A:1001747726820
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). Clustal-W - improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/Nar/22.22.4673
van der Geize, R., and Dijkhuizen, L. (2004). Harnessing the catabolic diversity of Rhodococci for environmental and biotechnological applications. Curr. Opin. Microbiol. 7, 255–261. doi: 10.1016/j.mib.2004.04.001
van der Geize, R., Hessels, G. I., van Gerwen, R., van der Meijden, R., and Dijkhuizen, L. (2002). Molecular and functional characterization of kshA and kshB, encoding two components of 3-ketosteroid 1-alpha-hydroxylase, a class IA monooxygenase, in Rhodococcus erythropolis strain SQ1. Mol. Microbiol. 45, 1007–1018. doi: 10.1046/j.1365-2958.2002.03069.x
van der Geize, R., Hessels, G. I., van Gerwen, R., Vrijbloed, J. W., van der Meijden, P., and Dijkhuizen, L. (2000). Targeted disruption of the kstD gene encoding a 3-ketosteroid delta(1)-dehydrogenase isoenzyme of Rhodococcus erythropolis strain SQ1. Appl. Environ. Microbiol. 66, 2029–2036. doi: 10.1128/Aem.66.5.2029-2036.2000
Winn, M. D., Ballard, C. C., Cowtan, K. D., Dodson, E. J., Emsley, P., Evans, P. R., et al. (2011). Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. 67, 235–242. doi: 10.1107/S0907444910045749
Yam, K. C., Geize, R., and Eltis, L. D. (2010). “Catabolism of aromatic compounds and steroids by Rhodococcus,” in Biology of Rhodococcus, ed. M. H. Alvarez (Berlin: Springer), 133–169.
Keywords: expression strain, flavoprotein, monooxygenase, phospholipid, Rhodococcus
Citation: Montersino S, te Poele E, Orru R, Westphal AH, Barendregt A, Heck AJR, van der Geize R, Dijkhuizen L, Mattevi A and van Berkel WJH (2017) 3-Hydroxybenzoate 6-Hydroxylase from Rhodococcus jostii RHA1 Contains a Phosphatidylinositol Cofactor. Front. Microbiol. 8:1110. doi: 10.3389/fmicb.2017.01110
Received: 20 April 2017; Accepted: 31 May 2017;
Published: 16 June 2017.
Edited by:
Daniela De Biase, Sapienza Università di Roma, ItalyReviewed by:
Hans-Peter Kohler, Swiss Federal Institute of Aquatic Science and Technology, SwitzerlandAlberto A. Iglesias, National University of the Littoral, Argentina
Copyright © 2017 Montersino, te Poele, Orru, Westphal, Barendregt, Heck, van der Geize, Dijkhuizen, Mattevi and van Berkel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Willem J. H. van Berkel, willem.vanberkel@wur.nl
†Present address: Stefania Montersino, Adienne Pharma & Biotech, Milan, Italy; Evelien te Poele and Lubbert Dijkhuizen, CarbExplore Research BV, Groningen, Netherlands; Roberto Orru, Institute of Molecular Biology, Johannes Gutenberg University, Mainz, Germany; Robert van der Geize, Laboratory of Pathology East-Netherlands, Hengelo, Netherlands
‡These authors have contributed equally to this work.
 Stefania Montersino1†‡
Stefania Montersino1†‡ Roberto Orru
Roberto Orru Adrie H. Westphal
Adrie H. Westphal Albert J. R. Heck
Albert J. R. Heck Willem J. H. van Berkel
Willem J. H. van Berkel