- 1Plant Breeding, Wageningen University & Research, Wageningen, Netherlands
- 2Biotechnology and Plant Breeding Department, Faculty of Agriculture, Ferdowsi University of Mashhad, Mashhad, Iran
There is currently limited knowledge on the role of hormones in plants responses to combinations of abiotic and pathogen stress factors. This study focused on the response of tomato near-isogenic lines (NILs) that carry the Ol-1, ol-2, and Ol-4 loci, conferring resistance to tomato powdery mildew (PM) caused by Oidium neolycopersici, to combined PM and salt stress. These NILs were crossed with the notabilis (ABA-deficient), defenceless1 (JA-deficient), and epinastic (ET overproducer) tomato mutants to investigate possible roles of hormone signaling in response to combined stresses. In the NILs, marker genes for hormonal pathways showed differential expression patterns upon PM infection. The epinastic mutation resulted in breakdown of resistance in NIL-Ol-1 and NIL-ol-2. This was accompanied by reduced callose deposition, and was more pronounced under combined salt stress. The notabilis mutation resulted in H2O2 overproduction and reduced susceptibility to PM in NIL-Ol-1 under combined stress, but lead to higher plant growth reduction under salinity and combined stress. Resistance in NIL-ol-2 was compromised by the notabilis mutation, which was potentially caused by reduction of callose deposition. Under combined stress the compromised resistance in NIL-ol-2 was restored. PM resistance in NIL-Ol-4 remained robust across all mutant and treatment combinations. Hormone signaling is critical to the response to combined stress and PM, in terms of resistance and plant fitness. ABA appears to be at the crossroads of disease susceptibility/senescence and plant performance under combined stress These gained insights can aid in narrowing down targets for improving crop performance under stress combinations.
Introduction
Plant hormones are central to plant adaptation to changing environmental conditions as well as to interactions with pathogenic and non-pathogenic organisms. To maximize fitness under different stress scenarios, resource allocation must be precisely prioritized, and to achieve that, hormonal signaling pathways are delicately interconnected and inter-regulated (Denance et al., 2013). Understanding the underlying regulatory mechanisms of hormone crosstalk is increasingly important in view of the current global climate change, which is projected to further intensify unfavorable conditions for crop production (Lobell et al., 2011; Challinor et al., 2014; Trnka et al., 2014). A significant consequence of climate change is the increased frequency of stress combinations that plants are exposed to, especially of abiotic factors in combination with pathogenic microorganisms (Garrett et al., 2006; Kissoudis et al., 2014; Suzuki et al., 2014).
Significant progress has been made in understanding hormone cross-regulation under individual stress factors, such as abiotic stress or defence response to pathogens. Abscisic acid (ABA) is the major orchestrator of adaptation and tolerance to abiotic stress (Yoshida et al., 2014), while interplay between salicylic acid (SA), jasmonic acid (JA), and ethylene (ET) regulates resistance responses to pathogens and pests (Pieterse et al., 2012). In the model plant Arabidopsis thaliana, SA is the main player in responses to biotrophic pathogens, while JA and ET, antagonistically with SA, mount defence against necrotrophs (Robert-Seilaniantz et al., 2011). This distinction is rather blurry in many occasions, as interactions between hormonal pathways depend on hormone concentration and timing of induction (Koornneef et al., 2008; Pieterse et al., 2009), and species-specific responses may be distinct from those reported in Arabidopsis. For example, in barley activation of systemic acquired resistance is under the control of ERF and WRKY transcription factors, but not of SA (Dey et al., 2014). In tomato, however, SA enhances resistance against necrotrophic pathogens such as Botrytis, while increasing susceptibility against biotrophs (Achuo et al., 2004).
With regard to interaction between abiotic and biotic stresses, a mostly antagonistic interaction was observed between ABA and defence signaling across many plant species. ABA negatively interacts with both SA and JA/ET signaling, compromising local and systemic acquired resistance to pathogens (Anderson et al., 2004; Yasuda et al., 2008; Ulferts et al., 2015). Increased disease resistance in ABA-deficient mutants strengthen this notion, though in many cases the increased resistance might be due to pleiotropic effects of ABA depletion (Curvers et al., 2010; Mang et al., 2012; Sanchez-Vallet et al., 2012). On the contrary, there are also evidence for positive roles of ABA signaling especially in pre-penetration defence responses through priming for cell wall fortifications and callose deposition (Ton et al., 2009; Garcia-Andrade et al., 2011). Thus, even though the majority of the studies indicate a negative role of ABA in defence responses, this does not preclude a potential beneficial contribution in specific pathosystems or at specific stages during pathogenesis.
Recent studies suggested non-additive interactions between responses to abiotic and biotic stresses at both phenotypic and molecular levels (Prasch and Sonnewald, 2013; Rasmussen et al., 2013; Kissoudis et al., 2014). The complexity of interactions under combined abiotic and biotic stress is further emphasized by the differential regulation of a significant number of SA, JA, and ET responsive genes under abiotic stress (Walia et al., 2007; Huang et al., 2008). How the up-regulation of defence signaling pathways under combined stress affects adaptation to abiotic stress has not been established yet, although there is evidence suggesting that up-regulation of SA signaling dampens ABA-mediated responses (Kim et al., 2011).
Our research is focused on the regulation of tomato resistance responses to the combination of salt stress and powdery mildew (PM) caused by Oidium neolycopersici. We previously demonstrated that PM resistance was negatively affected by 100 mM NaCl (EC level of 10 dS/m) in an introgression line population segregating for partial PM resistance (Kissoudis et al., 2015). Further research indicated that salt stress has the highest impact on disease susceptibility under mild stress conditions (EC∼6 dS/m, 50 mM NaCl), while more severe salinity restored resistance. Combined stress impacted plant performance significantly more than the individual stresses, which was manifested by accelerated senescence and leaf abscission. However, this response was strongly conditioned by the type of resistance to PM, as indicated by the examination of near-isogenic lines (NILs) carrying the resistance loci Ol-1, ol-2, and Ol-4 (Kissoudis et al., 2016). The dominant locus Ol-1 enhances basal defence by inducing delayed cell death (DCD) in the late stages of pathogen infection (Li et al., 2007; Seifi, 2011). The recessive gene ol-2, which is a mutant of the tomato MLO gene encoding a membrane protein, mediates resistance to PM by callose deposition and cell wall fortification to stop PM at the pre-penetration stage (Bai et al., 2008). The dominant locus Ol-4 is an NBS-LRR gene homologous to Mi-1 (Seifi et al., 2011), and triggers a hypersensitivity response (HR) to prevent the PM colonization after formation of primary haustoria (Bai et al., 2003; Li et al., 2006). Expression analysis of selected pathway marker genes indicated a significant role of ET and JA, which were uniquely highly upregulated in the tomato susceptible genotypes under combined stress (Kissoudis et al., 2016).
Here, we evaluated the effects of three major hormonal pathways, ABA, JA, and ET on different tomato resistance mechanisms to PM conferred by different Ol-genes (Ol-1, ol-2, and Ol-4). Two complementary strategies were adopted in this work. First, we monitored the expression of marker genes for different hormone pathways by using NILs that carry each of the different Ol-genes. Then, we evaluated whether PM resistance in these NILs is compromised under single (either salt or PM) or combined stress (salt and PM) when JA, ET, and ABA pathways are perturbated, by analyzing progenies of crosses between these NILs and ABA, JA, and ET hormone mutants. Our results provide a better understanding of how major hormonal pathways can affect tomato resistance and plant performance under combined PM and salt stress.
Materials and Methods
Plant and Fungal Materials
The recessive epinastic (epi), and notabilis (not) tomato mutants and their respective backgrounds AC (Ailsa Craig), and VNF8, were obtained from the Tomato Genetic Resource Center (TGRC), University of California, Davis, CA, USA. The tomato defenseless1 (def1) recessive mutant was obtained from Dr. C.A. Ryan, Washington State University. The near-isogenic lines NIL-Ol-1, NIL-ol-2, and NIL-Ol-4 [in the background of S. lycopersicum cv Moneymaker (MM)] confer monogenic resistance to PM through different mechanisms (Bai et al., 2005). Each of the NILs was crossed with the epi, not, and def1 mutants, with the exception of the NIL-Ol-4 cross with the not mutant. By subsequent selfing, homozygous progenies for both the Ol-gene and mutations were selected in the F3 and F4 generations, and were used for evaluation of response to combined PM and salt stress.
The pathogenic fungus O. neolycopersici originated from infected commercial tomato plants and was maintained on MM plants in a greenhouse compartment at 20 ± 3°C with 70 ± 15% relative humidity (RH).
Selection for the Presence of Ol-Genes and Hormonal Mutations
Selection for homozygous Ol-genes was carried out by using gene-based or tightly linked molecular markers for the resistance genes (Bai et al., 2005). The primers used for genotyping were: F-TGCTCTAACAAAATCACCAAAATC and R-AAATGGTCAAACAAAGTCTATTGAG for Ol-1, F-ACCCTTAAGAAATAGGGCAAA and R- ACCATCATGAACCCATGTCT for ol-2, and: F-GAACCGGATGTGTCCTTGAC and R-TTCTCCGAGACTTTGAACAAGA for Ol-4.
DNA isolation was carried out according to Wang et al. (1993) with some modifications. About 10 mg of leaf tissue was homogenized in 20 μl of 0.5 N NaOH for 5 min. Then 20 μl of 100 mM Tris-HCl was added and thoroughly mixed, and 5 μl of this homogenate was diluted with 95 μl of 100 mM Tris-HCl. The PCR reaction mix contained 0.12 μl Phire Hot Start II DNA Polymerase (Thermo Scientific), 2 μl forward primer (5 μM), 2 μl reverse primer (5 μM), 1 μl of the diluted leaf homogenate as a DNA template and 1 μl of PVP (10% w/v) as a chelating agent for impurities, into a final volume of 11 μl. The amplification profile was 40 cycles of 98°C for 5 s, 54°C for 5 s, and 72°C for 10 s.
Different selection approaches were used to select homozygous plants for hormonal mutation, depending on whether the gene and the polymorphism underlying the mutation are known. The not mutation is well-characterized and is the consequence of a specific A/T base pair deletion in the coding sequence resulting in a frameshift mutation (Burbidge et al., 1999), indicating that it is a null mutant. Homozygous plants for the not mutation were selected based on sequencing a PCR fragment harboring the A/T mutation at position 597 of the ORF (primers used: not-F: GTTCGAAACGGAGCTAACCC, not-R: AACAAGTCCGAAGAGCCCA). The gene mutation causing the epi phenotype is not known, but mutant seedlings carrying the epi mutation are significantly shorter that wild ones when germinated in the dark (Barry et al., 2001). Accordingly, seeds were germinated in the dark and plants showing no etiolation were selected as homozygous plants carrying the epi mutation. Selection for the def1 mutant was done based on the fact that this mutation affects JA biosynthesis (Howe et al., 1996). To test this, single leaflets of the wild type (WT), JA deficient parental lines were pierced and the induction of expression of the JA marker leucine aminopeptidase A (LapA) was monitored 24 h after wounding with qRT-PCR using primers F-ATCTCAGGTTTCCTGGTGGAAGGA, R-AGTTGCTATGGCAGAGGCAGAG. RNA isolation was performed with a MagMAXTM-96 Total RNA Isolation Kit in a KingFisherTM Flex Magnetic Particle Processor according to manufacturer’s instructions, and expression of the LapA gene was evaluated following the method described in the gene expression section bellow. An average of 100-fold difference in expression was observed between WT plants and the homozygous def1 mutant, indicating that LapA marker gene could be used safely as a qualitative marker for selecting the def1 mutantion.
Experimental Conditions and Treatments
Experiments were carried out in the greenhouse with a photoperiod regime of 16 h light and 8 h dark, and 70% RH. Additional lighting (100 Wm-2) was supplied if the incoming radiation was below 200 Wm-2. Plants were grown in pots filled with vermiculite irrigated with half strength Hoagland nutrient solution at regular intervals till leaching of the solution, in order to avoid accumulation of nutrients and NaCl.
The experiments were carried out twice in two different years (2013 and 2014) in the period of April–May. In the first experiment, the response of progenies of NILs × mutant crosses (named Ol-1 × def, ol-2 × def, etc.) that were homozygote for both the Ol gene and the mutation, or only homozygote for the Ol gene without the mutation, to PM was evaluated under normal and salt stress conditions For each combination, 6–8 plants were tested. Three-week-old plants were watered with half strength Hoagland solution containing either zero (no salt stress) or 50 mM NaCl (mild salt stress). Eight days after the initiation of salt stress treatments, plants were inoculated with PM by uniformly spraying a suspension of 5 × 104 conidia per ml prepared by washing conidial spores from leaves of heavily infected (sporulation stage) MM plants. Plants were grown for another 20 days after inoculation.
In the second experiment, all NIL × mutant crosses were similarly assessed, excluding the crosses that carried only the Ol-genes and not the hormonal mutations. In addition, we included a non-PM treatment (only salt stress): half of the plants from Ol-1 × epi or Ol-1 × not (selected based on their explicit phenotypes) were spatially isolated 8 days after the salt treatments and were not sprayed with PM. The plants thus were exposed to all possible treatment combinations (no salt stress/not inoculated, no salt stress/inoculated, salt stress/not inoculated, salt stress/inoculated). Plants were grown for another 20 days after the inoculation.
Plant Performance Evaluation under Salt Stress and PM
The disease severity was assessed at 10, 15, and 25 days post-inoculation (dpi) for the first experiment, and at 15 dpi for the second experiment, as disease index (DI) on a scale from 0 (no PM symptom) to 5 (heavy PM infection) as described before (Kissoudis et al., 2014). In addition to DI, a measure of senescence development [senescence index (SI)] was introduced to describe the accelerated senescence phenotypes observed at the late stages of infection under salt stress: 0 = healthy plant, 1 = 0.1–10% of foliar area affected, 2 = 10–20% area affected with yellowing and moderate wilting, 3 = 20–30% area affected with severe wilting, 4 = 30–50% area affected with severe wilting and moderate leaf abscission, and 5 = > 50% area affected with severe wilting and leaf abscission.
Ion Content Analysis
The five youngest leaves (from the top of the plant) were sampled at 20 dpi, the endpoint of the second experiment, in order to examine differences in actively growing tissues, potentially linked to growth performance, and avoid the severely senescing bottom leaves of susceptible genotypes under combined stress conditions. The concentration of Na+, Cl-, K+, PO43-, SO42-, Mg2+, and Ca2+ was measured with ion chromatography as described previously (Kissoudis et al., 2015).
In situ Histological Analyses of H2O2 Accumulation and Callose Deposition
Leaf disks (1.3 cm in diameter) were taken from leaflets of the 4th leaf counting from the bottom on the 3rd day after pathogen inoculation. To ensure uniformity, leaf disks were taken from the middle of the leaflets on both sides of the central vein. Staining was carried out in 24-well plates, where leaf disks were placed with the abaxial side up. For H2O2 visualization, leaf disks were stained in 1 mg/ml DAB (3-3′-diaminobenzidine), pH 3.7 for 16 h in the dark and were subsequently transferred to 96% ethanol for 24 h to remove chlorophyll (Martinez de Ilarduya et al., 2003). Leaf disks were mounted on glass slides with 70% glycerol.
Callose deposition visualization was performed according to the described method (Ton et al., 2005; Luna et al., 2011) with slight modifications. Leaf disks were initially placed in 96% ethanol to remove chlorophyll and after a 1-min wash in 0.07 M K2HPO4 (pH 9), were stained for 2 h in 0.02% (w/v) aniline blue in 0.07 M K2HPO4 (pH 9) at room temperature. Leaf disks were mounted on glass slides with 70% glycerol. Callose deposit was quantified from digital photographs by the number of white pixels (fluorescence, related to callose intensity) relative to the total number of pixels covering plant material using the software Adobe® Photoshop® CS6.
Gene Expression Analysis and Pathogen Quantification
For the time course expression study on hormonal marker genes in the NILs challenged only with PM, the same time series as used previously for cDNA-AFLP profiling (Li et al., 2007) were used in this experiment. In brief, this time series included cDNAs from plants of MM, NIL-Ol-1, NIL-ol-2 and NIL-Ol-4, and non-inoculated (mock) and PM-inoculated leaves (2nd and 3rd) of three plants per line were collected at 1, 3, 5, 7, and 9 dpi. For each line, the cDNAs from mock samples from different time points were pooled and used as calibrator for qRT-PCR analysis.
To evaluate the expression of stress, defence, and hormone marker genes under salt and PM stresses, leaflets from the 3rd and 4th leaves (counting from the bottom) were sampled at 6 dpi, when pathogen mycelium growth was not yet visible. Sampling for pathogen quantification was carried out at 14 dpi, when pathogen growth from the primary infection had reached its peak using leaflets from the 4th and 5th leaf counting from the bottom. For each genotype, 4–5 plants were used.
The RNA for gene expression analyses was isolated with the RNeasy Plant Mini Kit (Qiagen). The isolated RNA was treated with DNase I (Invitrogen) to remove residual DNA. cDNA synthesis was performed using 1 μg RNA template by iScriptTM cDNA Synthesis Kit (BioRAD). qRT-PCR was conducted using the iQ SYBR Green supermix (Bio-Rad) and the CFX96 Real-Time system (Bio-Rad). The reaction mix contained 5 μl 2× iQ SYBR GREEN super mix, 1 μl forward primer (3 μM), 1 μl reverse primer (3 μM), and 3 μl cDNA (or DNA, 20 ng) template, in a final volume of 10 μl. Thermocycling condition was 95°C for 3 min, followed by 40 cycles of 95°C for 30 s and 60°C for 30 s. Primers for tomato elongation factor 1α (EF) were Fw-EF-GGAACTTGAGAAGGAGCCTAAG and Rv-EF-CAACACCAACAGCAACAGTCT (Gao et al., 2014). The primers used to monitor the expression of tomato genes are described in Supplementary Table S1. Relative expression was calculated using the 2-ΔΔCt method (Livak and Schmittgen, 2001). Plant and fungal DNA for pathogen quantification analysis was extracted with DNeasy Plant Mini Kit (Qiagen). Primers used for fungal quantification were Fw-On-CGCCAAAGACCTAACCAAAA and Rv-On-AGCCAAGAGATCCGTTGTTG.
Results
Expression Pattern of Marker Genes for Hormonal Pathways in NILs
To monitor changes in JA, SA, ET, and ABA pathways, the expression level of marker genes for these pathways were measured in the NILs and MM, in a time course from 1 to 9 days after inoculation with PM. Significant differences were observed in the expression patterns and in the magnitude of induction for some of these marker genes in the NILs and MM (Figure 1).
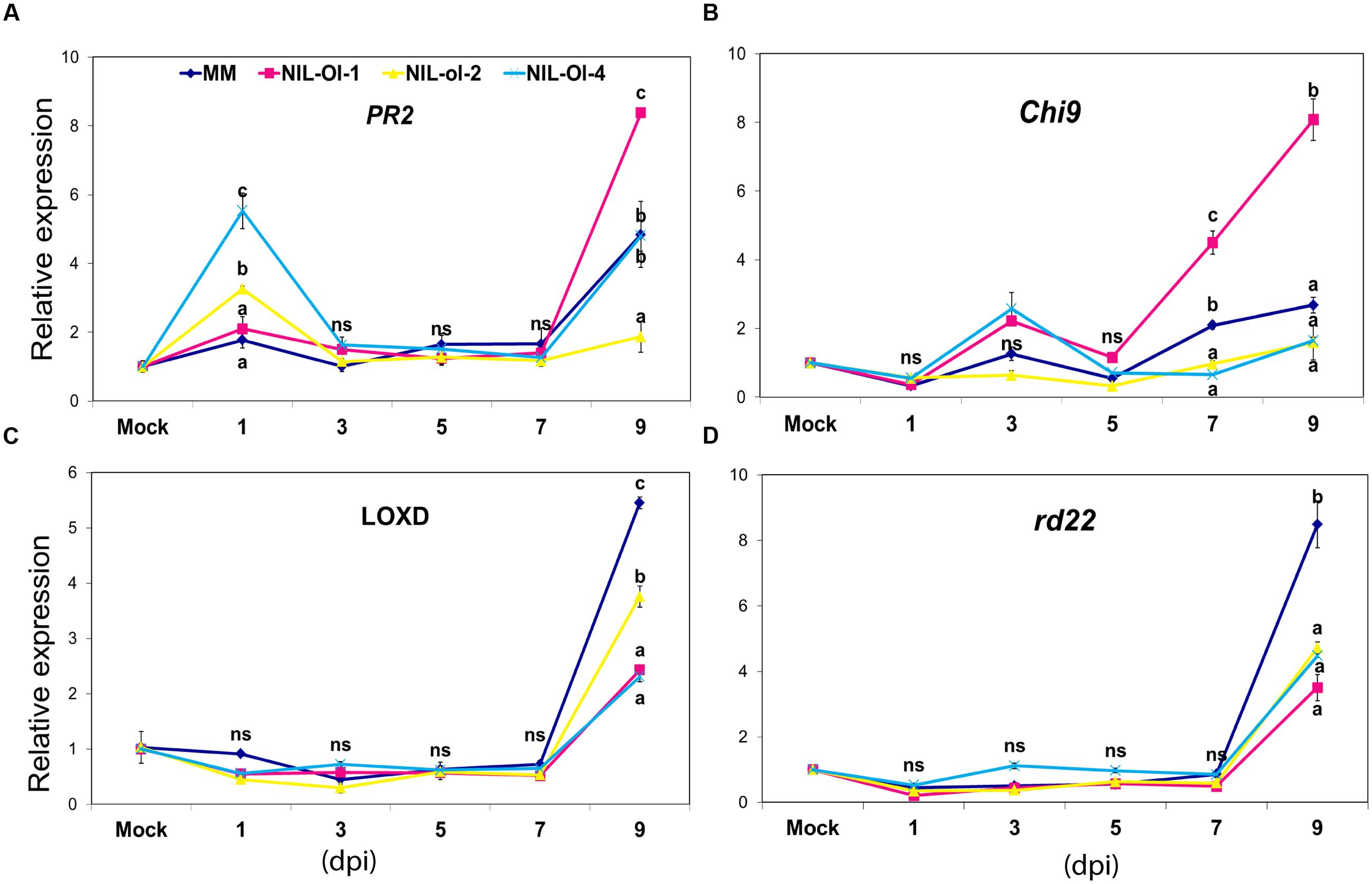
FIGURE 1. Expression of (A) PR-2, (B) rd22, (C) Chitinase9, and (D) LOXD (markers for SA, ABA, ET and JA and pathways, respectively) in MM, and NIL-Ol-1, -ol-2, and -Ol-4 in a time-course after inoculation with PM. Second and third leaves were sampled at 1, 3, 5, 7, and 9 days post-inoculation (dpi) from powdery mildew (PM)-inoculated and -non-inoculated (mock) plants. Statistically significant differences (P ≤ 0.05) between genotypes (within each timepoint of measurement) are designated with different letters. Error bars represent standard error of mean, (n = 3); ns, non-significant:
Salicylic acid induces expression of a group of pathogenesis-related genes (PR genes) in Arabidopsis including PR-2, which is often used as a marker gene for the SA pathway (Uknes et al., 1992). The tomato PR-2 gene (Domingo et al., 1994) was induced in response to Phytophthora infestans as well as in response to benzothiadiazole (BTH, an analog of SA; Beyer et al., 2001). Therefore, we used PR-2 as a marker gene for the SA pathway in this study (see SlPR-2 in Supplementary Table S1). At 1 dpi, there was an induction in the PR-2 expression in NIL-ol-2 and NIL-Ol-4, but very little induction in NIL-Ol-1 and MM. At the last time point (9 dpi), the PR-2 expression was increased in MM, NIL-Ol-1, and NIL-Ol-4, with the highest level in NIL-Ol-1 (approximately ninefold induction compared to non-inoculated plants).
The ET pathway signaling was monitored through the expression of the Chitinase9 (Chi9) gene, which has been used as a marker gene for ET pathway in tomato (Barry et al., 2001). The expression level of Chi9 did not show great fluctuations across genotypes and time points with the exception of NIL-Ol-1, in which a marked up-regulation was observed in the later time points (4.5- and 8-fold induction at 7 and 9 dpi, respectively).
The lipoxygenase D (LOXD) gene has been shown to be induced by JA in tomato (Heitz et al., 1997), thus we used this gene as a marker for the JA pathway. Its expression was relatively stable or slightly down-regulated across all genotypes till 7 dpi, but a marked up-regulation was observed in all genotypes at 9 dpi, which was strongest in MM and NIL-ol-2 (increase of approximately six and fourfold, respectively, compared to control conditions).
The Arabidopsis rd22 gene is ABA-responsive (Shinozaki et al., 2003). By performing TBLASTN in tomato Unigene database1 a homologue of this gene in tomato (EU679376.1) was retrieved and used as the tomato rd22 orthologue. Similar to the JA marker LOXD, rd22 expression was relatively stable among genotypes in 1–7 dpi, and was significantly up-regulated at 9 dpi. MM showed the highest expression, 8.5-fold increase compared to control conditions, while all NILs exhibited a fourfold upregulation.
Effects of Hormonal Mutants on the PM Resistance in the NILs under Combined Stresses
The mutants and their background lines, as well as their progeny that were homozygous for individual Ol-genes but do not carry the hormone mutations (null segregants) were evaluated for PM susceptibility. The hormone mutants and their background lines were all susceptible to PM, with a susceptibility level similar to that in MM. The null segregants, however, were as resistant as the NILs, suggesting that the resistance conferred by the Ol-genes was not affected by the genetic background in the crosses (data not shown).
Similar to our previous results, mild salt stress significantly increased the PM susceptibility of MM and NIL-Ol-1, while the resistance level of NIL-ol-2 and NIL-Ol-4 was not affected. When combined with hormone mutants, the resistance conferred by Ol-1 and ol-2 was significantly affected, but the resistance conferred Ol-4 was not (Figures 2A–D).
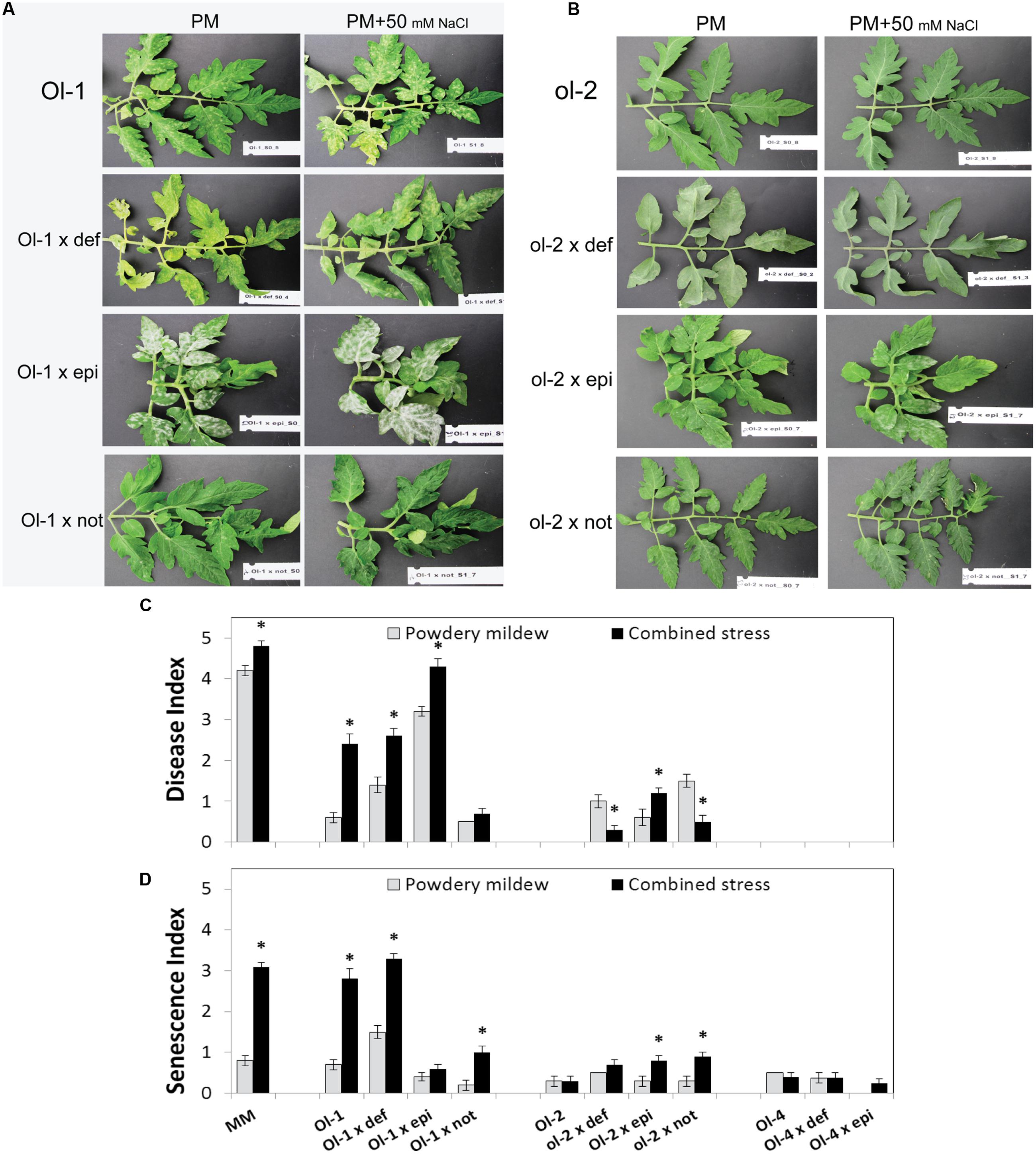
FIGURE 2. Leaf phenotypes of (A) NIL-Ol-1 and (B) NIL-ol-2 and their respective mutants under PM and combined stress. (C) Disease and (D) senescence index of NIL-O-1, -ol-2, -Ol-4, the recurrent parent MM and their crosses with different hormone mutants under PM individually (no salt stress) and in combination with 50 mM NaCl measured at 15 dpi. Error bars depict standard error (n = 6). Epi, epinastic (ET overproducer); not, notabilis (ABA deficient); def, defenseless1 (JA deficient). Asterisks denote statistically significant pairwise differences (P ≤ 0.05) between PM and combined stress (PM with salt) treatments for each genotype. Error bars represent standard error of mean.
Resistance conferred by the Ol-1 gene was compromised in plants carrying the epi mutation (e.g., average DI of 3.2 for Ol-1 × epi compared to 0.6 for NIL-Ol-1), with susceptibility increasing even further under salt stress (DI of 4.3 with salt combination compared to 3.2 without salt, Figure 2C, Supplementary Table S2). The significant increase in susceptibility of the Ol-1 × epi plants was accompanied by almost complete abolishment of the accelerated senescence and cell death symptoms observed in NIL-Ol-1 under salt stress (SI = 0.6 for Ol-1 × epi compared to SI = 2.8 for NIL-Ol-1, Figure 2D). The Ol-1 × not plants showed a level of resistance similar to NIL-Ol-1 plants without salt stress. However, ABA deficiency markedly suppressed the susceptibility response of Ol-1 under salt stress (DI = 0.7 for Ol-1 × not compared to DI = 2.4 for NIL-Ol-1 under combined stress, Figure 2C), and additionally reduced the accelerated senescence and leaf abscission phenotype (SI = 1 for Ol-1 × not compared to SI = 2.8 for NIL-Ol-1, Figure 2D). JA deficiency impacted senescence in PM treated Ol-1 × def plants with increased yellowing and abscission of older leaves (Supplementary Figure S1).
For the ol-2-mediated resistance, increased susceptibility was observed in ol-2 × def, ol-2 × epi, and ol-2 × not plants. Under salt stress, this susceptibility was significantly further increased for ol-2 × epi plants (DI = 1.2 with salt compared to DI = 0.8 without salt), while it was significantly decreased for ol-2 × def and ol-2 × not plants (DI = 0.3 and 0.5 with salt compared to DI = 1 and 1.5, respectively, without salt, Figure 2C, Supplementary Table S2). Slightly higher senescence was observed for ol-2 × epi and ol-2 × not under combined stress in comparison to NIL-ol-2 (Figure 2D).
Powdery mildew quantification by qPCR was in line with the visual scoring. In many occasions even greater differences between genotypes or treatments were observed. Only for NIL-Ol-1 plants and Ol-1 × def plants under combined stress the qPCR results revealed a smaller difference compared to what our visual scoring suggested, potentially due to the senescence symptoms leading to an overestimation of the visual disease score (Figure 3).
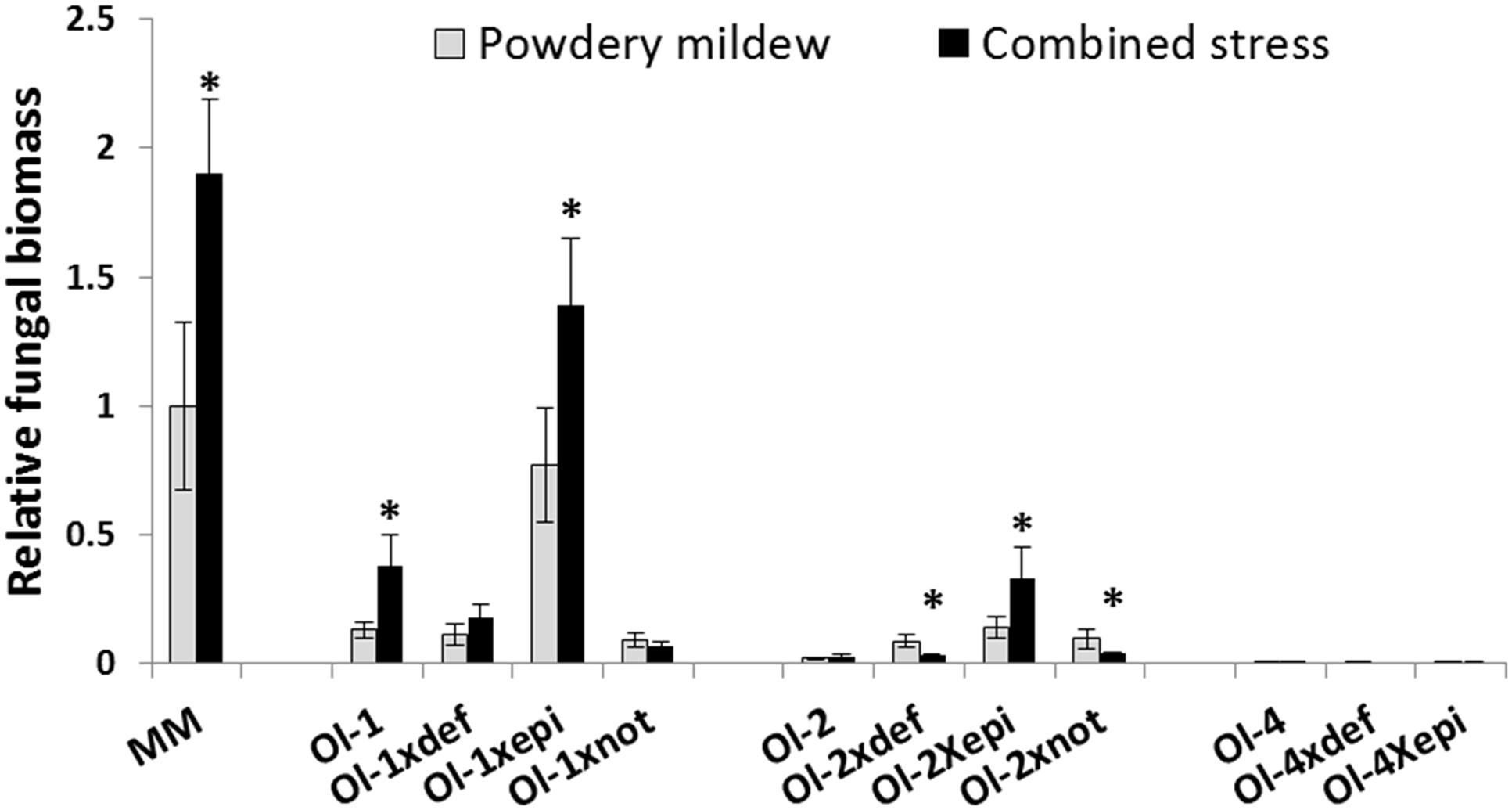
FIGURE 3. Relative Oidium neolycopersici fungal biomass [calculated as the ratio of fungal ITS gene amplification in comparison with tomato EF1a and normalized with the values of MM under PM infection (no salt stress)] in MM and NIL-Ol-1, -ol-2, and -Ol-4 and their respective mutants under PM infection alone and in combination with 50 mM NaCl. Asterisks denote statistically significant pairwise differences (P ≤ 0.05) between PM and combined stress (PM with salt) treatments for each genotype. Error bars represent standard error of mean (n = 4).
Performance and Fitness Cost of epi and not Mutations in NIL-Ol-1 and NIL-ol-2 under Combined Stress
Explicit phenotypes were observed for Ol-1 × epi, Ol-1 × not, ol-2 × epi, ol-2 × not plants. These lines were studied in more detail under control conditions, salt stress (50 mM), PM inoculation, and combined salt stress and PM inoculation, allowing an assessment of growth performance costs under the different stress conditions. These Ol-gene and mutant combinations are particularly interesting as ABA is the major hormone orchestrating abiotic stress responses in plants (Yoshida et al., 2014), while ET signaling was shown to be crucial for plant susceptibility and senescence responses under combined stress (Kissoudis et al., 2016).
The Ol-1 × epi and ol-2 × epi plants had reduced biomass under conditions without stress compared to the corresponding NIL lines, but exhibited higher relative stress tolerance (biomass under salt stress relative to biomass under control conditions). In contrast and as expected, ABA deficiency conferred by the not mutation further reduced biomass under salt stress relative to the control conditions (Figure 4). PM resulted in a decrease in aboveground fresh weight in all Ol-gene × mutant combinations. The combination of salt and PM imposed an even greater growth penalty than salt stress alone. While the reduction in performance caused by PM under combined salt stress relative to salt stress alone was lower in the not mutant crosses, the growth reduction resulting from the salt stress per se was far greater in these ABA-deficient plants than in any of the other tested plants (Figure 4).
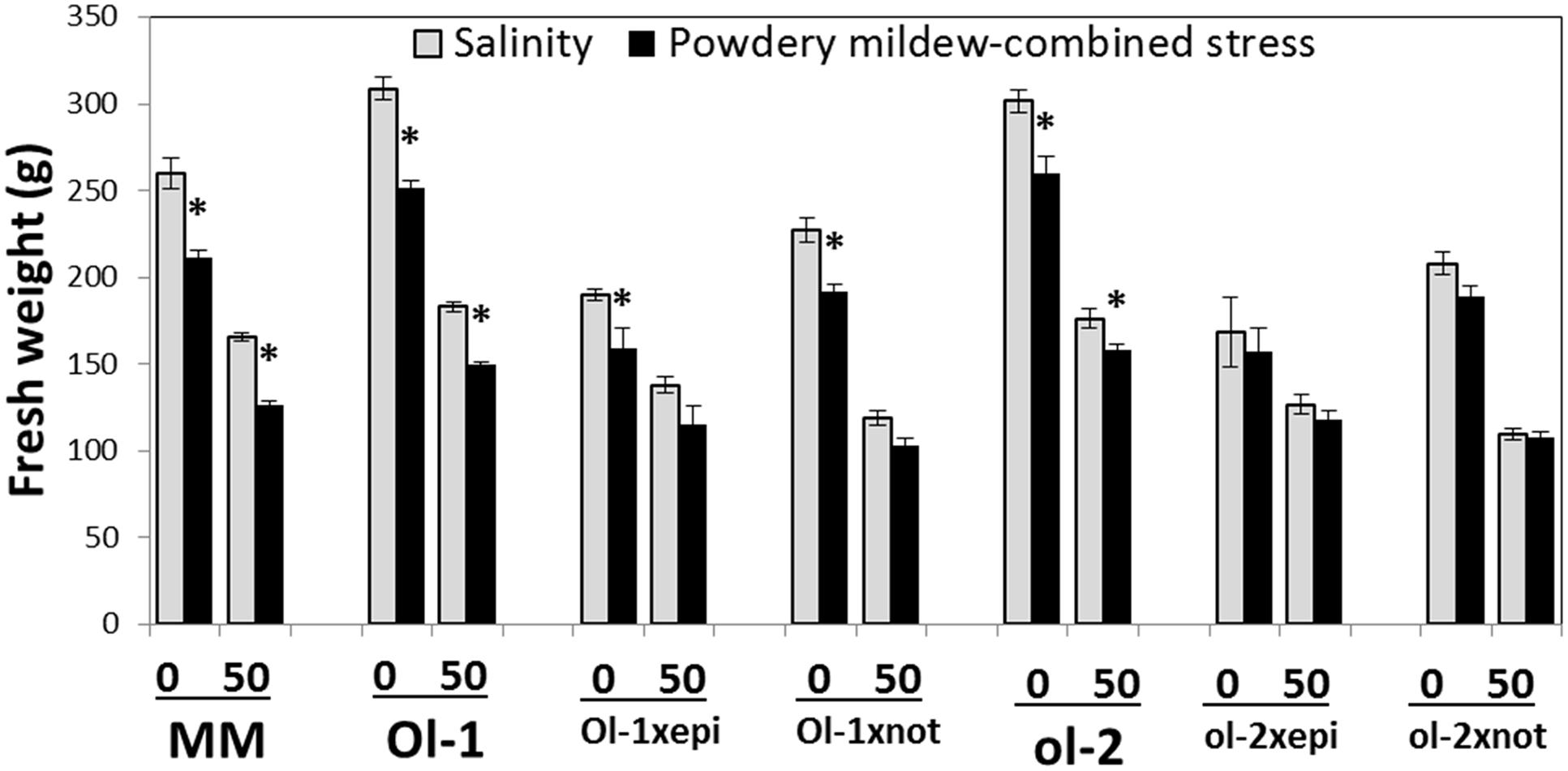
FIGURE 4. Aboveground biomass (FW) of MM, NIL-O-1, -ol-2, and their crosses with the hormone mutants under control conditions (0) and salt stress (50 mM NaCl) on the x-axis, and with or without PM (black vs. light gray). Level 0 for salinity stress corresponds to stress-free control conditions, while level 0 for PM-combined stress corresponds to PM infection alone (no salt stress). Asterisks denote statistically significant pairwise differences (P ≤ 0.05) between control/salinity and PM/combined stress for each genotype. Error bars represent standard error of mean (n = 4).
Ion content, and especially Na+ and Cl- concentration was shown to impact PM susceptibility (Kissoudis et al., 2016). Both the epi and not crosses with NIL-Ol-1 and NIL-ol-2 accumulated a higher amount of Na+ and Cl- under salt stress compared to the parental NILs (Supplementary Figure S2). However, the Ol-1 × epi and ol-2 × epi plants exhibited a significant reduction in the concentration of Na+ and Cl- under combined PM and salt stress compared to salt stress only, while the crosses with not mutant had slightly increased Na+ and Cl- concentrations under these conditions. K+ content was higher in Ol-1 × epi and ol-2 × epi plants in most of the treatments.
Histological Analysis of Callose Deposition and H2O2 Accumulation
Callose deposition at the attempted sites of pathogen penetration increases plant resistance and is a major hallmark of ol-2-mediated resistance (Bai et al., 2008; Ellinger et al., 2013). As shown previously (Kissoudis et al., 2016), NIL-ol-2 exhibited increased callose deposits compared to NIL-Ol-1 and MM upon PM infection, and additional salt stress decreased callose deposits in all genotypes (Figure 5). Under only PM infection, the epi mutation resulted in a near complete absence of callose deposits in the crosses with Ol-1 and ol-2. The not mutation did not significantly affect callose deposits in Ol-1 × not plants, but led to a threefold decrease of callose deposits in ol-2 × not plants (Figure 5, only PM infection). However, under combined stress, Ol-1 × not and especially ol-2 × not exhibited denser callose deposits compared with PM only.
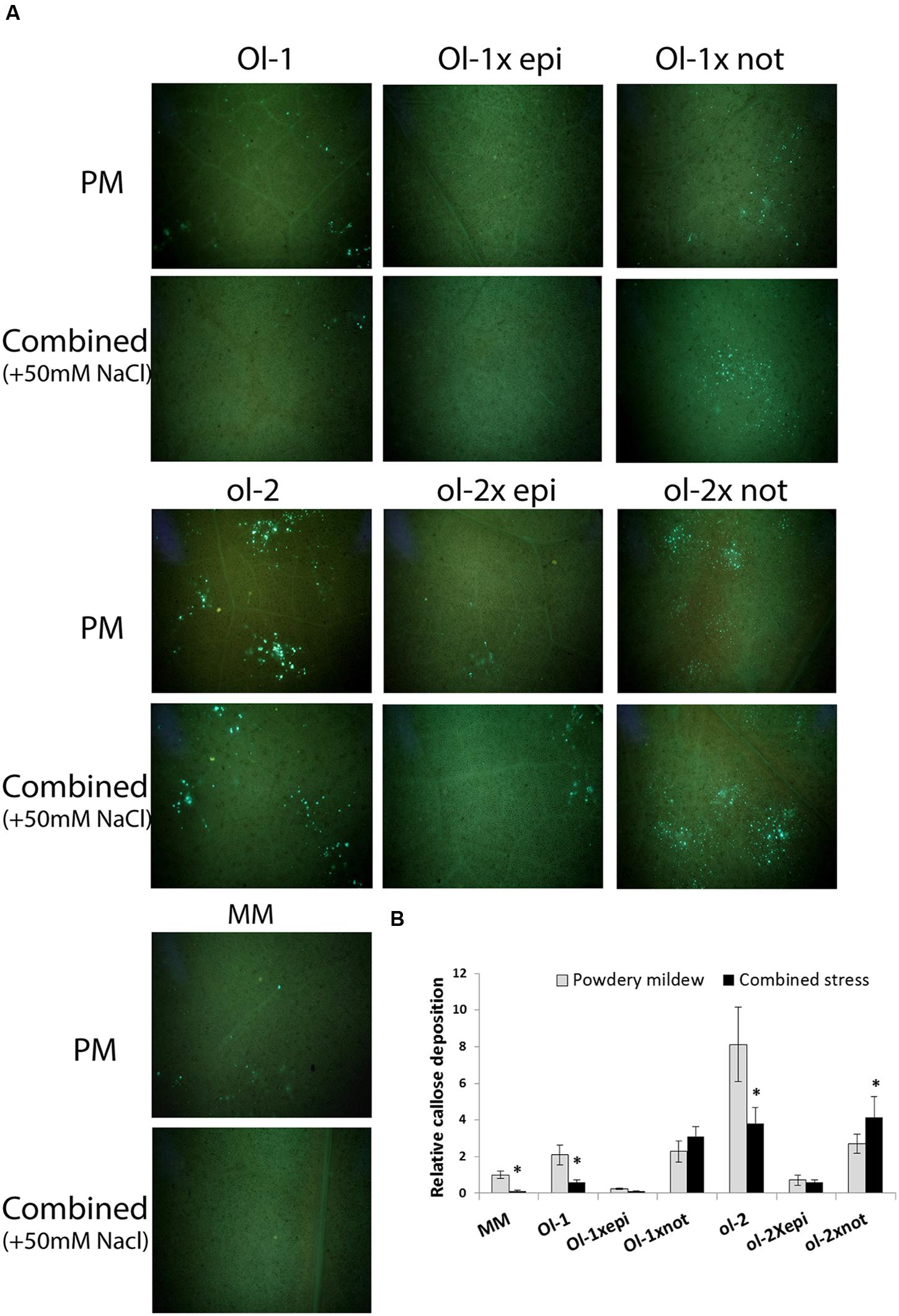
FIGURE 5. (A) Callose deposits in leaves of MM, NIL-Ol-1 and ol-2 and their respective crosses with epi and not mutants as visualized with UV microscopy after aniline blue staining, (B) quantification of callose deposition relative to MM under PM infection. Asterisks denote statistically significant pairwise differences (P ≤ 0.05) between PM and combined stress (PM with salt) treatments for each genotype. Error bars represent standard error of mean.
Examination of H2O2 content with DAB staining indicated slightly higher ROS production in MM, NIL-Ol-1, and NIL-ol-2 under individual stress (PM infection or salt stress) or combined stresses, compared to control plants of each genotype (Figure 6). The epi mutation did not influence H2O2 accumulation in the NILs. However, a massive H2O2 increase was observed in both Ol-1 × not and ol-2 × not plants.
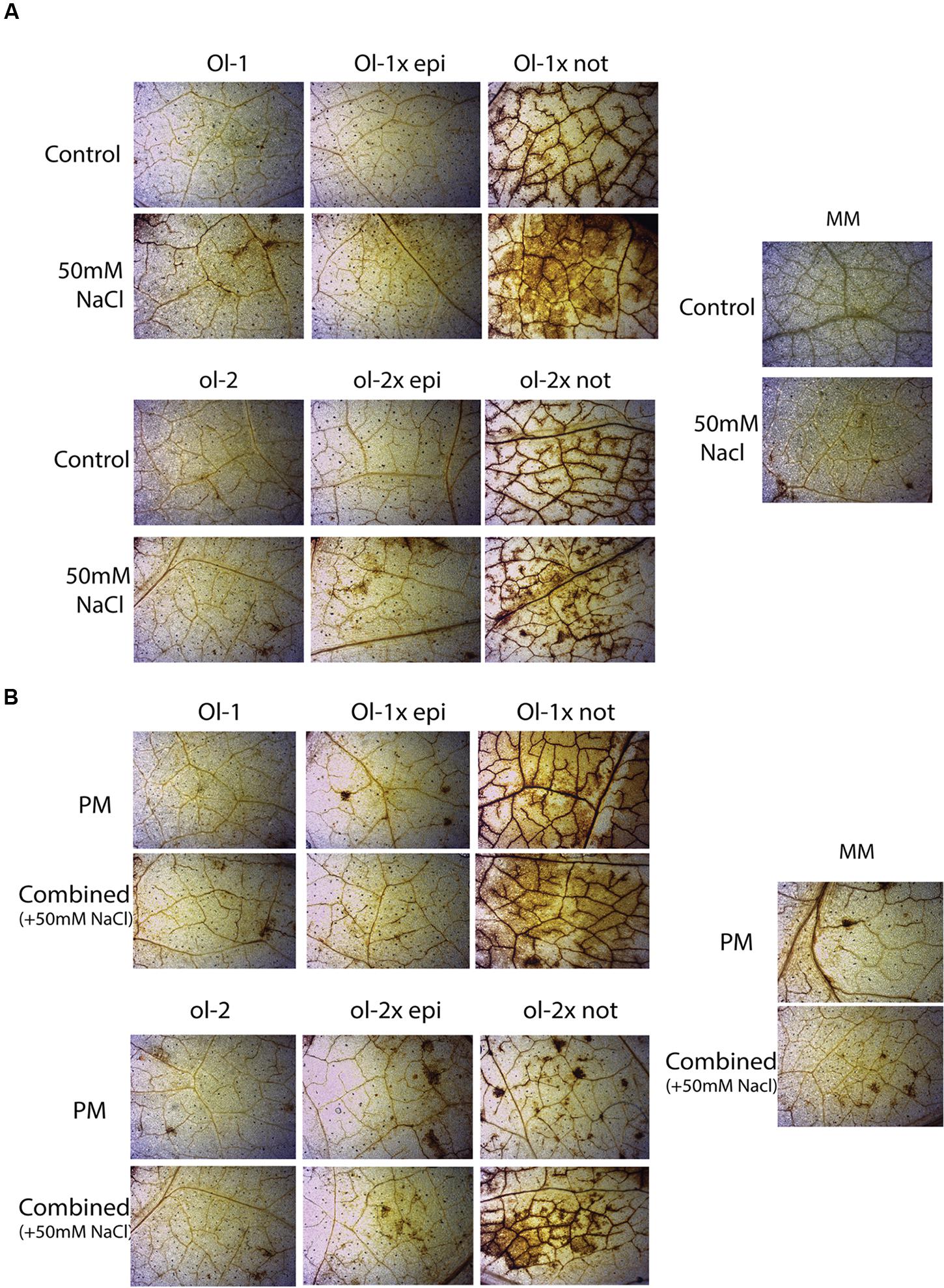
FIGURE 6. H2O2 visualization after DAB staining in MM, NIL-Ol-1 and ol-2 and their respective crosses with epi and not mutants under (A) control and salt stress, (B) PM and combined stress.
Expression Analyses
Gene expression of additional marker genes for the biosynthesis and signaling of major hormonal pathways, ROS, antioxidant, and ion homeostasis pathways involved in abiotic and biotic stress responses of tomato was monitored at 6 dpi, prior to visible PM symptoms (Figure 7, Supplementary Figure S3). The expression of the ABA biosynthesis gene NCED was either reduced (in Ol-1 × epi plants) or stable (in ol-2 × epi plants) under combined stress compared to salt stress only. In the not mutant this gene contains a mutation that causes a frameshift mutation. It may be transcribed but does not code for a functional enzyme. ABA deficiency in not is in line with the modest expression levels (significantly lower compared to NILs) of the ABA catabolic gene, ABAOH, and the dehydrin gene, DHN-TAS under all conditions. Dehydrin expression was highly induced in Ol-1 × epi and o1-2 × epi plants under salt stress (50- and 6-fold, respectively), but this response was completely abolished under combined stress, while it was exceptionally induced under combined stress in NIL-ol-2.
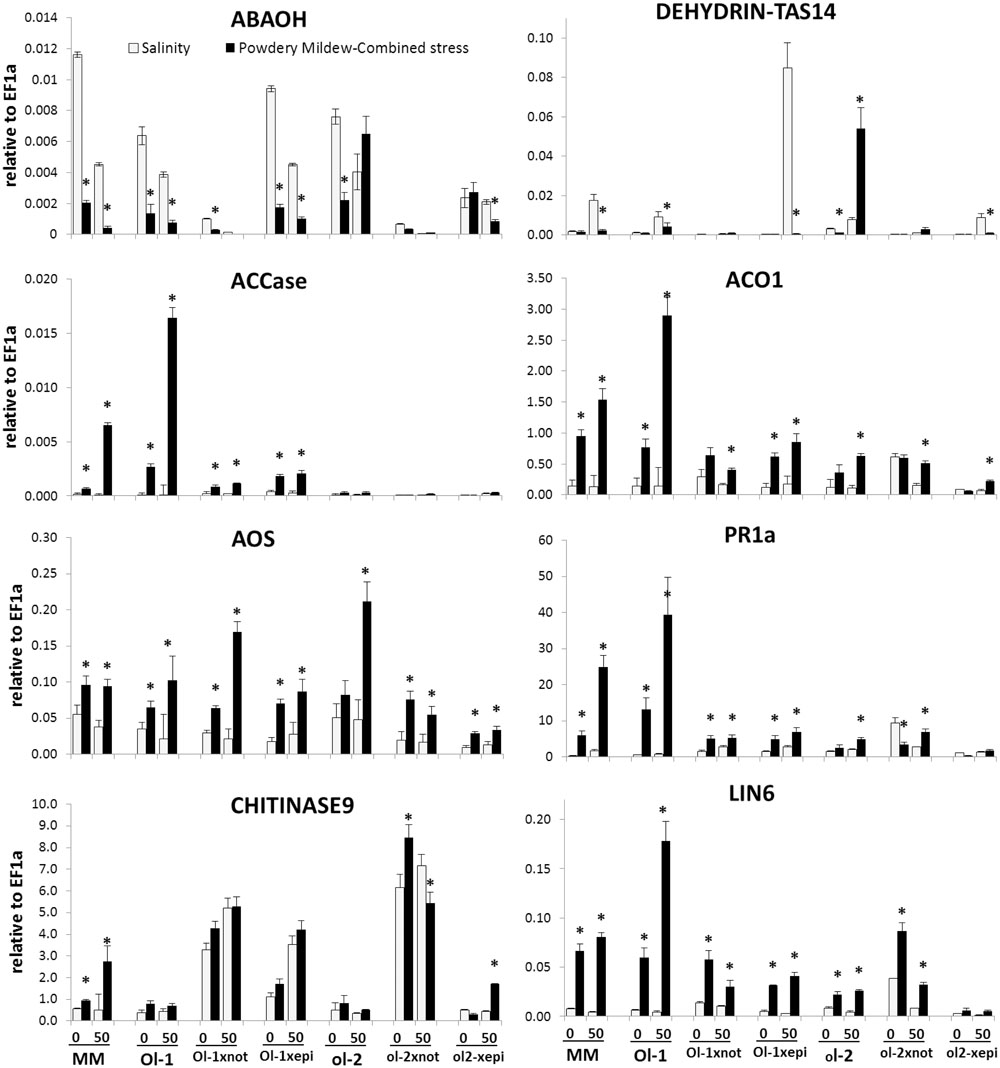
FIGURE 7. Expression of gene-markers for hormonal, abiotic, and biotic stress signaling pathways in MM, NIL-Ol-1 and ol-2 and their respective crosses with epi and not mutants, relative to EF1a, which was used as a housekeeping gene. Treatment and labeling scheme are the same as Figure 4. Asterisks denote statistically significant pairwise differences (P ≤ 0.05) between control/salinity and PM/combined stress for each genotype. Error bars represent standard error of mean (n = 4).
Under combined stress, an induction of ET biosynthetic genes ACCase and ACCox (ACO1) was observed in NIL-Ol-1, accompanying the increased PM susceptibility and senescence response. This induction was significantly reduced in Ol-1 × epi and Ol-1 × not plants. On the other hand, CHI9 was vastly induced in both Ol-1 × not and ol-2 × not plants (up to 20-fold compared to NIL-Ol-1 and NIL-ol-2) and these expression levels were maintained under all stress treatments.
Expression levels of AOS and LOXD genes, nodes of the JA pathway, were significantly reduced in ol-2 × epi and ol-2 × not crosses under salt and combined stress treatments (reductions up to sixfold). PR1a induction in NIL-Ol-1 after PM and combined stress (25- and 70-fold higher, respectively, compared to non-stress conditions) was greatly reduced in both Ol-1 × epi and Ol-1 × not plants, despite the higher basal expression in these plants. The strong induction of invertase LIN6 observed in NIL-Ol-1 under combined stress was greatly reduced in Ol-1 × epi and Ol-1 × not plants.
Discussion
The epi Mutation Compromises the Ol-1- and ol-2-Mediated PM Resistance
The Ol-1 gene confers incomplete PM resistance by inducing DCD upon PM infection (Li et al., 2007; Seifi, 2011). The gene is not cloned yet, but it is likely a non NBS-LRR gene, enhancing basal defence. The ol-2 gene is a mlo-mutant and mediates resistance to PM by callose deposition to stop PM at penetration stage (Bai et al., 2005). The PM resistance mediated by the Ol-1 and ol-2 genes was compromised by the epi mutation that is reported to produce high levels of ET (Fujino et al., 1988). This is in line with the established role of ET in susceptibility for biotrophic pathogens, in Arabidopsis and several other plant species, through negative interaction with SA signaling (Pieterse et al., 2009). ET signaling is also reported to be involved in disease symptom development caused by Xanthomonas campestris pv. vesicatoria in tomato (O’Donnell et al., 2003). Salt stress had an additional negative effect on the PM susceptibility of Ol-1 × epi and ol-2 × epi plants, indicating additive effects of this abiotic stress and ET in compromising Ol-1- and ol-2-mediated resistance. Salinity stress was shown to increase ET concentration in tomato (Amjad et al., 2014).
The overproduction of ET in the tomato epi mutant (Fujino et al., 1988) was reflected by the 10-fold induction of ACCase expression compared to WT (relative expression of 0.000049 in NIL-Ol-1 compared to 0.000372 in Ol-1 × epi, p < 0.001). No differences were observed in ACO1 expression, the final enzyme in ET biosynthesis (relative expression of 0.140 in NIL-Ol-1 compared to 0.126 in Ol-1 × epi, p = 0.263). This is in accordance with the previous study that showed ACC (the product of ACCase and the rate-limiting compound for ET synthesis) is increased in the epi mutant (Fujino et al., 1988).
The epi is a mutant with pleiotropic phenotypic effects such as reduced growth and leaf epinasty (Supplementary Figure S1) (Fujino et al., 1988; Barry et al., 2001). There is a striking difference between the increased senescence and cell death observed in NIL-Ol-1, and the complete absence of senescence and cell death in Ol-1 × epi plants under combined stress, which seems to contradict the known promotion of senescence by ET (Penfold and Buchanan-Wollaston, 2014). ET overproduction is also shown to stimulate ROS production and the accompanying symptoms (Bartoli et al., 2013), but this was not observed in the epi plants. In NIL-Ol-1, increased susceptibility under combined stress was accompanied by an induction in the expression of ET biosynthesis and responsive genes ACC and ACO1, but not for CHI9. In Ol-1 × epi plants, this increase was only modest for ACC and ACO1, which might indicate that ET biosynthesis may not have exceeded a threshold level that would impact senescence. Similar observations on the lack of important ET-induced symptoms such as increased senescence were reported previously for this mutant (Barry et al., 2001). Additional pleiotropic alterations at the cellular level were observed for epi by others, especially changes in the epidermal cells which differ from the WT by having a more round-shaped and swollen cells (Barry et al., 2001). These changes can be functionally significant for biotic stress responses through a potential effect on the cytoskeleton dynamics and the secretion and deposition of anti-fungal compounds. Manipulation of these processes resulted in a significant effect on exocytosis mechanisms, which are linked to the transport of antifungal compounds at the site of infection and increased susceptibility in PAMP-mediated resistance (PTI), but did not affect HR-mediated resistance (Hardham et al., 2007; Miklis et al., 2007; Henty-Ridilla et al., 2014). This is similar to our findings of reduced callose deposition in Ol-1 × epi and ol-2 × epi plants while the HR-based resistance conferred by Ol-4 was not affected by the epi mutation.
The not Mutation Differentially Affects PM Resistance Mediated by the Ol-1 and ol-2 Genes
The not mutation, which induces ABA deficiency (Mulholland et al., 2003), had both positive and negative impacts on disease resistance conferred by the Ol-genes. It slightly, but significantly, increased susceptibility of NIL-ol-2 after PM infection, while no significant changes were observed for NIL-Ol-1. Under combined stress, the increased susceptibility and senescence of NIL-Ol-1 was significantly alleviated in the Ol-1 × not plants (Figure 2). In ol-2 × not plants a slight decrease of susceptibility was also observed under combined stress. These results indicate a complex interaction between ABA signaling and disease resistance in alignment with a number of other studies (Audenaert et al., 2002; De Torres Zabala et al., 2009; Curvers et al., 2010; Mang et al., 2012), while the salt stress adds another layer of complexity. Both ROS production (increased) and callose deposition (decreased) were significantly affected in Ol-1 × not and ol-2 × not plants and might underlie the differential resistance responses of these plants.
A ROS-induced oxidative burst was shown to contribute to defense against Botrytis cinerea in the tomato ABA-deficient mutant sitiens (Asselbergh et al., 2007). However, recent findings indicate only a minimal effect of the ROS-induced oxidative burst on pathogenicity (Samalova et al., 2014). In ol-2 × not plants, the reduced callose deposition may have facilitated PM penetration, with enhanced growth of the pathogen overriding the impact of increased ROS levels. The addition of salt stress partially decreased disease symptoms in ol-2 × not plants, accompanied by increased callose deposition. The increased callose deposition under combined salt and PM stress may result from the partial restoration of ABA signaling by exposure to stress (Mulholland et al., 2003), positively affecting callose deposition. The not mutant has about 10–15% of the ABA levels of the WT (Mulholland et al., 2003). The addition of salt stress may have resulted in induction of additional ABA responsive tomato NCED genes; the ABA marker DHN-TAS was induced 10-fold.
The elevated levels of Na+ and Cl- concentration under combined stress might contribute to salinity-induced increased resistance, as a result of ion toxicity to the fungus. The levels observed in the not mutants at 50 mM NaCl were similar to the levels observed in MM plants under 150 mM NaCl, and this was shown to reduce disease progression (Kissoudis et al., 2016). Alternatively, the not-induced resistance may be linked to the unique increase in the expression of CHI9 (Figure 7), which is considered to be a component of ET signaling in tomato (Wu and Bradford, 2003) and has direct antifungal properties (Hong and Hwang, 2006).
The most pronounced effect of the not mutation under combined stress was the marked attenuation of senescence and leaf abscission in Ol-1 × not plants. This occurred despite of very high levels of ROS, which are known to be associated with senescence (Gregersen et al., 2013), although H2O2 alone was insufficient in triggering cell death in tobacco in response to bacteria (Mur et al., 2005). Our results indicate that ABA induces senescence under combined stress, with recent studies supporting this finding (Yang et al., 2014). Uncontrolled cell death and senescence under combined stress may therefore be mediated through the ABA signaling pathway. The reduced expression of ET biosynthesis and response genes in the not crosses with Ol-genes suggests that ET signaling regulation may be involved in this phenotypic response. Both synergistic and antagonistic regulation of ABA and ET have been described, but ABA appears to enhance ET levels under abiotic stress (Albacete et al., 2009), which is in agreement with our results. Further support for a role of ET in the combined stress-induced phenotype of the not crosses with Ol-genes is given by the fact that the ET level in not and sitiens ABA deficient mutants is lower compared to WT plants (Nitsch et al., 2012).
Hormonal Pathways Expression during PM Pathogenesis and Connections with the Phenotypes of NILs × Mutant Crosses
Ethylene signaling is induced in NIL-Ol-1 late during the time course analysis and the epinastic mutation might be disrupting this pattern resulting in increased susceptibility (Figure 1). Stress-induced ABA signaling may contribute to the observed susceptibility in NIL-Ol-1 as ABA signaling is also induced in the susceptible MM in response to PM infection. The restoration of the compromised Ol-1-conferred resistance in Ol-1 × not further supports a role of ABA for the compromised resistance of NIL-Ol-1 under combined stress.
Jasmonic acid signaling is induced in the resistant NIL-ol-2 challenged with PM, and disruption of JA signaling in the ol-2 × def cross results in partial breakdown of resistance. OPR3 silencing in tomato, which diminished OPDA and JA biosynthesis, resulted in reduced callose deposition (Scalschi et al., 2015), suggesting a connection between JA and callose deposition. Therefore, it is conceivable to draw a cause and effect link between JA-deficiency and lower callose deposition, and enhanced PM susceptibility in ol-2 × def. The reversal of susceptibility in ol-2 crosses with JA and ABA mutants under combined stress, suggests that abiotic stress may act synergistically with the mechanisms contributing to ol-2-mediated resistance.
Performance Costs and Benefits of epi and not Mutations in NIL-Ol-1 and NIL-ol-2 under Combined Stress
Despite the positive effect of decreasing senescence under combined stress, ABA deficiency had a severe plant performance cost in terms of fresh weight under salt and combined stress. The ABA pathway appears to underlie the antagonistic effects between abiotic and biotic stress (Yasuda et al., 2008; Sanchez-Vallet et al., 2012). Therefore ABA signaling should be studied in more details under combined stress, and include examination of downstream signaling components that enhance disease resistance without compromising abiotic stress adaptation and vice versa (Garcia-Andrade et al., 2011).
Although the epi mutant increased PM susceptibility, it resulted in a better growth under salt and combined stress. However, the growth penalty of the Ol-gene × epi plants under control conditions should be taken into account when considering the epi mutation for improving stress tolerance of commercially grown tomato under multiple stress conditions. Nevertheless, adapting ET signaling for improving crop resilience is an interesting strategy, which is supported by several studies identifying a positive contribution of ET signaling components in adaptation to abiotic stress (Cheng et al., 2013; Jiang et al., 2013; Peng et al., 2014).
ABA, JA, and ET Pathways Have no Influence on the Resistance Mediated by the Ol-4 Gene
In contrast to Ol-1 and ol-2, the resistance mediated by Ol-4 was stable under all treatments and with all hormone mutant combinations. Ol-4 is a homolog of the Mi-1 gene, encoding an NBS-LRR protein (Seifi et al., 2011). It triggers HR in a single epidermal cell in which fungal growth is stopped completely (Bai et al., 2003; Li et al., 2006). R-gene resistance is based on effector-triggered immunity (ETI), which is characterized by compensatory relationships between its different signaling components. The ETI response is stronger and more prolonged than PTI (Tsuda et al., 2009), making it more robust and less prone to negative regulation from environmental or genetic factors. Since resistance conferred by Ol-4 was not affected by the large genetic perturbations disrupting whole hormonal pathways in the hormonal mutants, it has the potential to be stable in combination with larger changes in hormone signaling pathways conferring abiotic stress tolerance. Whether only Ol-4 has this potential or it is applicable to R-genes in general remains to be established.
Conclusion
Ethylene appears to be central in the plant responses under combined stress, increasing PM susceptibility but promoting salt tolerance. The role of ABA and JA appears to be more complex, as their effect was dependent on the type of resistance and the co-occurrence of salt stress. ABA deficiency appears to limit senescence symptoms, but with significant trade-off between plant salt tolerance and growth. More detailed studies should be carried out to identify specific components of the ABA signaling pathway with fewer pleiotropic effects that can be more effectively implemented to increase combined stress tolerance in crops. Further research is required to delineate the synergistic and antagonistic relationships between signaling components under combined stress and to implement them in precision breeding approaches. Alternatively, the stacking of robust R-genes, like Ol-4, with well-established abiotic stress tolerance-conferring genes may provide robust resistance under combined abiotic and biotic stress.
Author Contributions
CK participated in the design of the study, performed the marker selection, phenotypic, histological, gene expression analyses and data analysis and wrote manuscript. AS and ZY generated the crosses and performed an initial phenotypic characterization. AI and HvdS performed the marker selection and phenotypic characterization. CvdW, RV, CvdL, and YB designed the study, supervised this work and assisted with editing the manuscript. All authors read and approved the final manuscript.
Funding
CK was supported by an “Alexander S. Onassis” Public Benefit Foundation scholarship. This research was made possible by a financial grant from the Dutch Topsector TKI Uitgangsmaterialen (project number 280).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
This work is part of the Ph.D. theses of the CK and AS (Seifi, 2011; Kissoudis, 2016).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.02009/full#supplementary-material
TABLE S1 | Primers used for expression analyses with qRT-PCR.
TABLE S2 | Multiple comparisons (protected LSD, P ≤ 0.05) of disease index and senescence under powdery mildew (PM) and combined stress (PM+Salt). Statistically significant differences between genotypes are designated with different letters.
FIGURE S1 | Whole plant phenotypes of NIL-Ol-1 and its crosses with mutants under powdery mildew (PM) and in combination with 50 Mm NaCl.
FIGURE S2 | Na+, Cl-, K+, SO42-, Mg2+, and Ca2+concentration in MM, NIL-Ol-1, and NIL-ol-2 and their respective crosses with epi and not. Treatment and labeling scheme are the same as Figure 3. Asterisks denote statistically significant differences (P ≤ 0.05) between salinity and PM-combined stress for individual genotypes.
FIGURE S3 | Expression of additional genes-markers of hormonal, abiotic, and biotic stress signaling pathways relative to EF1a, which was used as a housekeeping gene. Treatment and labeling scheme are the same as Figure 4.
Footnotes
References
Achuo, E. A., Audenaert, K., Meziane, H., and Hofte, M. (2004). The salicylic acid-dependent defence pathway is effective against different pathogens in tomato and tobacco. Plant Pathol. 53, 65–72. doi: 10.1046/j.1365-3059.2003.a00947.x
Albacete, A., Martinez-Andujar, C., Ghanem, M. E., Acosta, M., Sanchez-Bravo, J., Asins, M. J., et al. (2009). Rootstock-mediated changes in xylem ionic and hormonal status are correlated with delayed leaf senescence, and increased leaf area and crop productivity in salinized tomato. Plant Cell Environ. 32, 928–938. doi: 10.1111/j.1365-3040.2009.01973.x
Amjad, M., Akhtar, J., Anwar-ul-Haq, M., Yang, A., Akhtar, S. S., and Jacobsen, S.-E. (2014). Integrating role of ethylene and ABA in tomato plants adaptation to salt stress. Sci. Hortic. 172, 109–116. doi: 10.1016/j.scienta.2014.03.024
Anderson, J. P., Badruzsaufari, E., Schenk, P. M., Manners, J. M., Desmond, O. J., Ehlert, C., et al. (2004). Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell 16, 3460–3479. doi: 10.1105/tpc.104.025833
Asselbergh, B., Curvers, K., Franca, S. C., Audenaert, K., Vuylsteke, M., Van Breusegem, F., et al. (2007). Resistance to Botrytis cinerea in sitiens, an abscisic acid-deficient tomato mutant, involves timely production of hydrogen peroxide and cell wall modifications in the epidermis. Plant Physiol. 144, 1863–1877. doi: 10.1104/pp.107.099226
Audenaert, K., De Meyer, G. B., and Hofte, M. M. (2002). Abscisic acid determines basal susceptibility of tomato to Botrytis cinerea and suppresses salicylic acid-dependent signaling mechanisms. Plant Physiol. 128, 491–501. doi: 10.1104/pp.010605
Bai, Y., Huang, C., van der Hulst, R., Meijer-Dekens, F., Bonnema, G., and Lindhout, P. (2003). QTLs for tomato powdery mildew resistance (Oidium lycopersici) in Lycopersicon parviflorum G1. 1601 co-localize with two qualitative powdery mildew resistance genes. Mol. Plant Microbe Interact. 16, 169–176. doi: 10.1094/MPMI.2003.16.2.169
Bai, Y., Pavan, S., Zheng, Z., Zappel, N., Reinstädler, A., Lotti, C., et al. (2008). Naturally occurring broad-spectrum powdery mildew resistance in a Central American tomato accession is caused by loss of mlo function. Mol. Plant Microbe Interact. 21, 30–39. doi: 10.1094/MPMI-21-1-0030
Bai, Y., van der Hulst, R., Bonnema, G., Marcel, T. C., Meijer-Dekens, F., Niks, R. E., et al. (2005). Tomato defense to Oidium neolycopersici: dominant Ol genes confer isolate-dependent resistance via a different mechanism than recessive ol-2. Mol. Plant Microbe Interact. 18, 354–362. doi: 10.1094/MPMI-18-0354
Barry, C. S., Fox, E. A., Yen, H., Lee, S., Ying, T., Grierson, D., et al. (2001). Analysis of the ethylene response in the epinastic mutant of tomato. Plant Physiol. 127, 58–66. doi: 10.1104/pp.127.1.58
Bartoli, C. G., Casalongue, C. A., Simontacchi, M., Marquez-Garcia, B., and Foyer, C. H. (2013). Interactions between hormone and redox signalling pathways in the control of growth and cross tolerance to stress. Environ. Exp. Bot. 94, 73–88. doi: 10.1016/j.envexpbot.2012.05.003
Beyer, K., Binder, A., Boller, T., and Collinge, M. (2001). Identification of potato genes induced during colonization by Phytophthora infestans. Mol. Plant Pathol. 2, 125–134. doi: 10.1046/j.1364-3703.2001.00059.x
Burbidge, A., Grieve, T. M., Jackson, A., Thompson, A., McCarty, D. R., and Taylor, I. B. (1999). Characterization of the ABA-deficient tomato mutant notabilis and its relationship with maize Vp14. Plant J. 17, 427–431. doi: 10.1046/j.1365-313X.1999.00386.x
Challinor, A. J., Watson, J., Lobell, D. B., Howden, S. M., Smith, D. R., and Chhetri, N. (2014). A meta-analysis of crop yield under climate change and adaptation. Nat. Clim. Change 4, 287–291. doi: 10.1038/Nclimate2153
Cheng, M. C., Liao, P. M., Kuo, W. W., and Lin, T. P. (2013). The Arabidopsis ETHYLENE RESPONSE FACTOR1 regulates abiotic stress-responsive gene expression by binding to different cis-acting elements in response to different stress signals. Plant Physiol. 162, 1566–1582. doi: 10.1104/pp.113.221911
Curvers, K., Seifi, H., Mouille, G., de Rycke, R., Asselbergh, B., Van Hecke, A., et al. (2010). Abscisic acid deficiency causes changes in cuticle permeability and pectin composition that influence tomato resistance to Botrytis cinerea. Plant Physiol. 154, 847–860. doi: 10.1104/pp.110.158972
De Torres Zabala, M., Bennett, M. H., Truman, W. H., and Grant, M. R. (2009). Antagonism between salicylic and abscisic acid reflects early host-pathogen conflict and moulds plant defence responses. Plant J. 59, 375–386. doi: 10.1111/j.1365-313X.2009.03875.x
Denance, N., Sanchez-Vallet, A., Goffner, D., and Molina, A. (2013). Disease resistance or growth: the role of plant hormones in balancing immune responses and fitness costs. Front. Plant Sci. 4:155. doi: 10.3389/fpls.2013.00155
Dey, S., Wenig, M., Langen, G., Sharma, S., Kugler, K. G., Knappe, C., et al. (2014). Bacteria-triggered systemic immunity in barley is associated with WRKY and ETHYLENE RESPONSIVE FACTORs but not with salicylic acid. Plant Physiol. 166, 2133–2151. doi: 10.1104/pp.114.249276
Domingo, C., Conejero, V., and Vera, P. (1994). Genes encoding acidic and basic class-III beta-1,3-glucanases are expressed in tomato plants upon viroid infection. Plant Mol. Biol. 24, 725–732. doi: 10.1007/BF00029854
Ellinger, D., Naumann, M., Falter, C., Zwikowics, C., Jamrow, T., Manisseri, C., et al. (2013). Elevated early callose deposition results in complete penetration resistance to powdery mildew in Arabidopsis. Plant Physiol. 161, 1433–1444. doi: 10.1104/pp.112.211011
Fujino, D. W., Burger, D. W., Yang, S. F., and Bradford, K. J. (1988). Characterization of an Ethylene Overproducing Mutant of Tomato (Lycopersicon esculentum Mill. Cultivar VFN8). Plant Physiol. 88, 774–779. doi: 10.1104/pp.88.3.774
Gao, D., Huibers, R. P., Loonen, A. E., Visser, R. G., Wolters, A. M., and Bai, Y. (2014). Down-regulation of acetolactate synthase compromises Ol-1- mediated resistance to powdery mildew in tomato. BMC Plant Biol. 14:32. doi: 10.1186/1471-2229-14-32
Garcia-Andrade, J., Ramirez, V., Flors, V., and Vera, P. (2011). Arabidopsis ocp3 mutant reveals a mechanism linking ABA and JA to pathogen-induced callose deposition. Plant J. 67, 783–794. doi: 10.1111/j.1365-313X.2011.04633.x
Garrett, K. A., Dendy, S. P., Frank, E. E., Rouse, M. N., and Travers, S. E. (2006). Climate change effects on plant disease: genomes to ecosystems. Annu. Rev. Phytopathol. 44, 489–509. doi: 10.1146/annurev.phyto.44.070505.143420
Gregersen, P. L., Culetic, A., Boschian, L., and Krupinska, K. (2013). Plant senescence and crop productivity. Plant Mol. Biol. 82, 603–622. doi: 10.1007/s11103-013-0013-8
Hardham, A. R., Jones, D. A., and Takemoto, D. (2007). Cytoskeleton and cell wall function in penetration resistance. Curr. Opin. Plant Biol 10, 342–348. doi: 10.1016/j.pbi.2007.05.001
Heitz, T., Bergey, D. R., and Ryan, C. A. (1997). A gene encoding a chloroplast-targeted lipoxygenase in tomato leaves is transiently induced by wounding, systemin, and methyl jasmonate. Plant Physiol. 114, 1085–1093. doi: 10.1104/pp.114.3.1085
Henty-Ridilla, J. L., Li, J., Day, B., and Staiger, C. J. (2014). ACTIN DEPOLYMERIZING FACTOR4 regulates actin dynamics during innate immune signaling in Arabidopsis. Plant Cell 26, 340–352. doi: 10.1105/tpc.113.122499
Hong, J. K., and Hwang, B. K. (2006). Promoter activation of pepper class II basic chitinase gene, CAChi2, and enhanced bacterial disease resistance and osmotic stress tolerance in the CAChi2-overexpressing Arabidopsis. Planta 223, 433–448. doi: 10.1007/s00425-005-0099-6
Howe, G. A., Lightner, J., Browse, J., and Ryan, C. A. (1996). An octadecanoid pathway mutant (JL5) of tomato is compromised in signaling for defense against insect attack. Plant Cell 8, 2067–2077. doi: 10.1105/tpc.8.11.2067
Huang, D., Wu, W., Abrams, S. R., and Cutler, A. J. (2008). The relationship of drought-related gene expression in Arabidopsis thaliana to hormonal and environmental factors. J. Exp. Bot. 59, 2991–3007. doi: 10.1093/jxb/ern155
Jiang, C., Belfield, E. J., Cao, Y., Smith, J. A., and Harberd, N. P. (2013). An Arabidopsis soil-salinity-tolerance mutation confers ethylene-mediated enhancement of sodium/potassium homeostasis. Plant Cell 25, 3535–3552. doi: 10.1105/tpc.113.115659
Kim, T. H., Hauser, F., Ha, T., Xue, S., Bohmer, M., Nishimura, N., et al. (2011). Chemical genetics reveals negative regulation of abscisic acid signaling by a plant immune response pathway. Curr. Biol. 21, 990–997. doi: 10.1016/j.cub.2011.04.045
Kissoudis, C. (2016). Genetics and Regulation of Combined Abiotic and Biotic Stress Tolerance in Tomato. Ph.D. thesis, Wageningen University, Wageningen.
Kissoudis, C., Chowdhury, R., van Heusden, S., van de Wiel, C., Finkers, R., Visser, R. G. F., et al. (2015). Combined biotic and abiotic stress resistance in tomato. Euphytica 202, 317–332. doi: 10.1007/s10681-015-1363-x
Kissoudis, C., Sunarti, S., van de Wiel, C., Visser, R. G. F., van der Linden, C. G., and Bai, Y. (2016). Responses to combined abiotic and biotic stress in tomato are governed by stress intensity and resistance mechanism. J. Exp. Bot. 67, 5119–5132. doi: 10.1093/jxb/erw285
Kissoudis, C., van de Wiel, C., Visser, R. G. F., and Van Der Linden, G. (2014). Enhancing crop resilience to combined abiotic and biotic stress through the dissection of physiological and molecular crosstalk. Front. Plant Sci. 5:207. doi: 10.3389/fpls.2014.00207
Koornneef, A., Leon-Reyes, A., Ritsema, T., Verhage, A., Den Otter, F. C., Van Loon, L. C., et al. (2008). Kinetics of salicylate-mediated suppression of jasmonate signaling reveal a role for redox modulation. Plant Physiol. 147, 1358–1368. doi: 10.1104/pp.108.121392
Li, C., Bai, Y., Jacobsen, E., Visser, R., Lindhout, P., and Bonnema, G. (2006). Tomato defense to the powdery mildew fungus: differences in expression of genes in susceptible, monogenic-and polygenic resistance responses are mainly in timing. Plant Mol. Biol. 62, 127–140. doi: 10.1007/s11103-006-9008-z
Li, C., Bonnema, G., Che, D., Dong, L., Lindhout, P., Visser, R., et al. (2007). Biochemical and molecular mechanisms involved in monogenic resistance responses to tomato powdery mildew. Mol. Plant Microbe Interact. 20, 1161–1172. doi: 10.1094/MPMI-20-9-1161
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lobell, D. B., Schlenker, W., and Costa-Roberts, J. (2011). Climate trends and global crop production since 1980. Science 333, 616–620. doi: 10.1126/science.1204531
Luna, E., Pastor, V., Robert, J., Flors, V., Mauch-Mani, B., and Ton, J. (2011). Callose deposition: a multifaceted plant defense response. Mol. Plant Microbe Interact. 24, 183–193. doi: 10.1094/MPMI-07-10-0149
Mang, H. G., Qian, W. Q., Zhu, Y., Qian, J., Kang, H. G., Klessig, D. F., et al. (2012). Abscisic acid deficiency antagonizes high-temperature inhibition of disease resistance through enhancing nuclear accumulation of resistance proteins SNC1 and RPS4 in Arabidopsis. Plant Cell 24, 1271–1284. doi: 10.1105/tpc.112.096198
Martinez de Ilarduya, O., Xie, Q., and Kaloshian, I. (2003). Aphid-induced defense responses in Mi-1-mediated compatible and incompatible tomato interactions. Mol. Plant Microbe Interact. 16, 699–708. doi: 10.1094/MPMI.2003.16.8.699
Miklis, M., Consonni, C., Bhat, R. A., Lipka, V., Schulze-Lefert, P., and Panstruga, R. (2007). Barley MLO modulates actin-dependent and actin-independent antifungal defense pathways at the cell periphery. Plant Physiol. 144, 1132–1143. doi: 10.1104/pp.107.098897
Mulholland, B. J., Taylor, I. B., Jackson, A. C., and Thompson, A. J. (2003). Can ABA mediate responses of salinity stressed tomato. Environ. Exp. Bot. 50, 17–28. doi: 10.1016/S0098-8472(02)00110-7
Mur, L. A. J., Kenton, P., and Draper, J. (2005). In planta measurements of oxidative bursts elicited by avirulent and virulent bacterial pathogens suggests that H2O2 is insufficient to elicit cell death in tobacco. Plant Cell Environ. 28, 548–561. doi: 10.1111/j.1365-3040.2005.01301.x
Nitsch, L., Kohlen, W., Oplaat, C., Charnikhova, T., Cristescu, S., Michieli, P., et al. (2012). ABA-deficiency results in reduced plant and fruit size in tomato. J. Plant Physiol. 169, 878–883. doi: 10.1016/j.jplph.2012.02.004
O’Donnell, P. J., Schmelz, E., Block, A., Miersch, O., Wasternack, C., Jones, J. B., et al. (2003). Multiple hormones act sequentially to mediate a susceptible tomato pathogen defense response. Plant Physiol. 133, 1181–1189. doi: 10.1104/pp.103.030379
Penfold, C. A., and Buchanan-Wollaston, V. (2014). Modelling transcriptional networks in leaf senescence. J. Exp. Bot. 65, 3859–3873. doi: 10.1093/jxb/eru054
Peng, J., Li, Z., Wen, X., Li, W., Shi, H., Yang, L., et al. (2014). Salt-induced stabilization of EIN3/EIL1 confers salinity tolerance by deterring ROS accumulation in Arabidopsis. PLoS Genet. 10:e1004664. doi: 10.1371/journal.pgen.1004664
Pieterse, C. M., Leon-Reyes, A., Van der Ent, S., and Van Wees, S. C. (2009). Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 5, 308–316. doi: 10.1038/nchembio.164
Pieterse, C. M., Van der Does, D., Zamioudis, C., Leon-Reyes, A., and Van Wees, S. C. (2012). Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 28, 489–521. doi: 10.1146/annurev-cellbio-092910-154055
Prasch, C. M., and Sonnewald, U. (2013). Simultaneous application of heat, drought, and virus to Arabidopsis plants reveals significant shifts in signaling networks. Plant Physiol. 162, 1849–1866. doi: 10.1104/pp.113.221044
Rasmussen, S., Barah, P., Suarez-Rodriguez, M. C., Bressendorff, S., Friis, P., Costantino, P., et al. (2013). Transcriptome responses to combinations of stresses in Arabidopsis. Plant Physiol. 161, 1783–1794. doi: 10.1104/pp.112.210773
Robert-Seilaniantz, A., Grant, M., and Jones, J. D. (2011). Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annu. Rev. Phytopathol. 49, 317–343. doi: 10.1146/annurev-phyto-073009-114447
Samalova, M., Meyer, A. J., Gurr, S. J., and Fricker, M. D. (2014). Robust anti-oxidant defences in the rice blast fungus Magnaporthe oryzae confer tolerance to the host oxidative burst. New Phytol. 201, 556–573. doi: 10.1111/nph.12530
Sanchez-Vallet, A., Lopez, G., Ramos, B., Delgado-Cerezo, M., Riviere, M. P., Llorente, F., et al. (2012). Disruption of abscisic acid signaling constitutively activates Arabidopsis resistance to the necrotrophic fungus Plectosphaerella cucumerina. Plant Physiol. 160, 2109–2124. doi: 10.1104/pp.112.200154
Scalschi, L., Sanmartin, M., Camanes, G., Troncho, P., Sanchez-Serrano, J. J., Garcia-Agustin, P., et al. (2015). Silencing of OPR3 in tomato reveals the role of OPDA in callose deposition during the activation of defense responses against Botrytis cinerea. Plant J. 81, 304–315. doi: 10.1111/tpj.12728
Seifi, A. (2011). Characterization of Tomato Genes for Resistance to Oidium Neolycopersici. Wageningen: Wageningen University.
Seifi, A., Kaloshian, I., Vossen, J., Che, D., Bhattarai, K. K., Fan, J., et al. (2011). Linked, if not the same, Mi-1 homologues confer resistance to tomato powdery mildew and root-knot nematodes. Mol. Plant Microbe Interact. 24, 441–450. doi: 10.1094/MPMI-06-10-0145
Shinozaki, K., Yamaguchi-Shinozaki, K., and Seki, M. (2003). Regulatory network of gene expression in the drought and cold stress responses. Curr. Opin. Plant Biol. 6, 410–417. doi: 10.1016/S1369-5266(03)00092-X
Suzuki, N., Rivero, R. M., Shulaev, V., Blumwald, E., and Mittler, R. (2014). Abiotic and biotic stress combinations. New Phytol. 203, 32–43. doi: 10.1111/nph.12797
Ton, J., Flors, V., and Mauch-Mani, B. (2009). The multifaceted role of ABA in disease resistance. Trends Plant Sci. 14, 310–317. doi: 10.1016/j.tplants.2009.03.006
Ton, J., Jakab, G., Toquin, V., Flors, V., Iavicoli, A., Maeder, M. N., et al. (2005). Dissecting the beta-aminobutyric acid-induced priming phenomenon in Arabidopsis. Plant Cell 17, 987–999. doi: 10.1105/tpc.104.029728
Trnka, M., Rotter, R. P., Ruiz-Ramos, M., Kersebaum, K. C., Olesen, J. E., Zalud, Z., et al. (2014). Adverse weather conditions for European wheat production will become more frequent with climate change. Nat. Clim. Change 4, 637–643. doi: 10.1038/Nclimate2242
Tsuda, K., Sato, M., Stoddard, T., Glazebrook, J., and Katagiri, F. (2009). Network properties of robust immunity in plants. PLoS Genet. 5:e1000772. doi: 10.1371/journal.pgen.1000772
Uknes, S., Mauch-Mani, B., Moyer, M., Potter, S., Williams, S., Dincher, S., et al. (1992). Acquired resistance in Arabidopsis. Plant Cell 4, 645–656. doi: 10.1105/tpc.4.6.645
Ulferts, S., Delventhal, R., Splivallo, R., Karlovsky, P., and Schaffrath, U. (2015). Abscisic acid negatively interferes with basal defence of barley against Magnaporthe oryzae. BMC Plant Biol 15:7. doi: 10.1186/s12870-014-0409-x
Walia, H., Wilson, C., Condamine, P., Liu, X., Ismail, A. M., and Close, T. J. (2007). Large-scale expression profiling and physiological characterization of jasmonic acid-mediated adaptation of barley to salinity stress. Plant Cell Environ. 30, 410–421. doi: 10.1111/j.1365-3040.2006.01628.x
Wang, H., Qi, M., and Cutler, A. J. (1993). A simple method of preparing plant samples for PCR. Nucleic Acids Res. 21, 4153–4154. doi: 10.1093/nar/21.17.4153
Wu, C.-T., and Bradford, K. J. (2003). Class I Chitinase and β-1,3-Glucanase are differentially regulated by wounding, methyl jasmonate, ethylene, and gibberellin in tomato seeds and leaves. Plant Physiol. 133, 263–273. doi: 10.1104/pp.103.024687
Yang, J., Worley, E., and Udvardi, M. (2014). A NAP-AAO3 regulatory module promotes chlorophyll degradation via ABA biosynthesis in Arabidopsis leaves. Plant Cell 26, 4862–4874. doi: 10.1105/tpc.114.133769
Yasuda, M., Ishikawa, A., Jikumaru, Y., Seki, M., Umezawa, T., Asami, T., et al. (2008). Antagonistic interaction between systemic acquired resistance and the abscisic acid-mediated abiotic stress response in Arabidopsis. Plant Cell 20, 1678–1692. doi: 10.1105/tpc.107.054296
Keywords: abscisic acid, senescence, cell death, callose, ROS burst, chitinase
Citation: Kissoudis C, Seifi A, Yan Z, Islam ATMT, van der Schoot H, van de Wiel CCM, Visser RGF, van der Linden CG and Bai Y (2017) Ethylene and Abscisic Acid Signaling Pathways Differentially Influence Tomato Resistance to Combined Powdery Mildew and Salt Stress. Front. Plant Sci. 7:2009. doi: 10.3389/fpls.2016.02009
Received: 04 October 2016; Accepted: 19 December 2016;
Published: 09 January 2017.
Edited by:
Agata Gadaleta, University of Bari Aldo Moro, ItalyReviewed by:
Francisco Perez-Alfocea, Spanish National Research Council, SpainAna Carmen Cohen, National University of Cuyo, Argentina
Copyright © 2017 Kissoudis, Seifi, Yan, Islam, van der Schoot, van de Wiel, Visser, van der Linden and Bai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuling Bai, bai.yuling@wur.nl
 Christos Kissoudis
Christos Kissoudis Alireza Seifi
Alireza Seifi Zhe Yan1
Zhe Yan1 A. T. M. Tanjimul Islam
A. T. M. Tanjimul Islam Clemens C. M. van de Wiel
Clemens C. M. van de Wiel Richard G. F. Visser
Richard G. F. Visser C. G. van der Linden
C. G. van der Linden Yuling Bai
Yuling Bai