Abstract
Powdery mildew (PM, caused by Podosphaera fusca) and downy mildew (DM, caused by Pseudoperonospora cubensis) are important diseases of cucumber (Cucumis sativus). Breeding for resistance has been undertaken since the 1940s, but underlying resistance genes have not been functionally analysed yet. The published genome sequence of cucumber catalyses the search for such genes. Genetic studies have indicated that resistances to PM and DM in cucumber are often inherited recessively, which indicates the presence of susceptibility genes (S-genes). Therefore we analyzed the cucumber genome for homologs of functionally proven S-genes known from other plant species. We identified 13 MLO-like genes in cucumber, three of which cluster in Clade V, the clade that contains all known MLO-like susceptibility genes to powdery mildews in other dicots. The expression of one of these three genes, CsaMLO1, located on chromosome 1, was upregulated after PM inoculation. It co-localizes with a QTL for PM resistance previously identified. Also homologs of the susceptibility genes PMR4 and PMR5 are located at this QTL. The second MLO-like gene from Clade V (CsaMLO8) resides in a recessively inherited major QTL for PM resistance at the bottom of chromosome 5, together with a PMR6-like gene. Two major QTL for DM recessive resistance at the top of chromosome 5 co-localize with CsaDMR6-2, which is homologous to the DMR6 susceptibility gene in Arabidopsis. This study has identified several candidate genes for susceptibility to PM and DM in cucumber that may explain QTL for recessively inherited resistance, reported earlier.
Similar content being viewed by others
Introduction
Disease resistant crops are commonly bred by the introgression of resistance (R) genes from wild relatives. However, race-specific resistance conferred by R-genes asserts selective pressure on pathogen populations, therewith overcoming resistance. Also resistance to powdery mildew (PM) in cucumber (Podosphaera fusca (Fr.) Braun & Shishkoff, syn. Sphaerotheca fuliginea Schlech ex Fr. Poll) has been bred through introgression of race specific resistance genes, which led to emergence of virulent races (Cohen et al. 2004; Torés et al. 2009). Pyramiding of R-genes has been proposed as a solution to this problem. An alternative strategy for achieving durable resistance is disabling genes that are required for susceptibility, namely susceptibility genes (S-genes) (Pavan et al. 2009).
Several natural loss-of-function alleles of S-genes are known in agriculture, providing durable disease resistance. The most well-known examples are the barley mlo mutants (Acevedo-Garcia et al. 2014). These barley mutants have been successfully employed in European barley growing for more than 35 years (Lyngkjær et al. 2000), emphasizing the durability of mlo-mediated disease resistance under agricultural conditions. Another example is the loss-of-function mutation in the proline-containing protein Pi21 in rice, providing resistance to rice blast throughout a century of cultivation (Fukuoka et al. 2009).
After discovery of the MLO susceptibility gene in barley, MLO-like susceptibility genes have been discovered in other monocots, i.e. OsMLO3 in rice (Devoto et al. 2003), TaMLO_A1 and TaMLO_B1 in wheat (Devoto et al. 2003; Várallyay et al. 2012). But also in dicots MLO-like susceptibility genes have been discovered, such as AtMLO2, AtMLO6 and AtMLO12 in Arabidopsis (Consonni et al. 2006), SlMLO1 in tomato (Bai et al. 2008), PsMLO1 in pea (Humphry et al. 2011; Pavan et al. 2011; Santo et al. 2013), CaMLO2 in pepper (Kim and Hwang 2012; Zheng et al. 2013), LjMLO1 in lotus (Humphry et al. 2011), and MtMLO1 in barrel clover (Humphry et al. 2011). The MLO-genes have about seven transmembrane domains, and are located in the plasma membrane with an extracellular amino terminus and an intracellular carboxy terminus with a calmodulin binding domain (Kim et al. 2002). In spite of many efforts to elucidate the biochemical mechanism of the MLO-mediated susceptibility to PM, this still remains a mystery to a large extent (Acevedo-Garcia et al. 2014).
In addition to MLO other genes that are required for susceptibility to PM have been detected in Arabidopsis, i.e. PMR4 (powdery mildew resistance 4; Nishimura et al. 2003; Ellinger et al. 2013), PMR5 (Vogel et al. 2004) and PMR6 (Vogel et al. 2002). These three genes appear to influence the cell wall of the plant. Loss-of-function mutations in these PMR genes resulted in PM resistance in Arabidopsis. PMR4 encodes callose synthase (Nishimura et al. 2003). When PM infection occurred in Arabidopsis thaliana, the plant cells responded by depositing callose, a (1,3)-b-glucan polymer, at the penetration site, thus thickening the cell wall to block the fungal penetration (Nishimura et al. 2003). If view of this, PMR4 would encode a resistance gene (PM Resistance), rather than a susceptibility gene. Nishimura et al. (2003) provide a possible explanation for this paradox, as callose or callose synthase would negatively regulate the defence by the salicylic acid pathway. Ellinger et al. (2013) confirmed that overexpression of PMR4 in A. thaliana leads to callose deposition at PM penetration sites, but in their findings this leads to resistance, rather than to susceptibility. However, Huibers et al. (2013) showed that down-regulation of the PMR4 ortholog in tomato reduced the susceptibility to PM, indicating the S-gene effect of PMR4. PMR5 belongs to a large family of plant-specific genes with unknown function. The pmr5 mutant exhibited pectin enrichment and had smaller cells (Vogel et al. 2004). PMR6 encodes a pectate lyase–like protein (Vogel et al. 2002).
For downy mildew (DM), several S-genes were revealed in Arabidopsis, i.e. DMR1 (van Damme et al. 2009) and DMR6 (van Damme et al. 2008). The DMR1 gene encodes homoserine kinase. dmr1 mutants contained high levels of homoserine, that would trigger a novel form of DM resistance, independent of known immune responses (van Damme et al. 2009). Interestingly, the tomato ortholog of DMR1 was found to be required for PM susceptibility in this crop (Huibers et al. 2013). DMR6 encodes a putative 2OG-Fe(II) oxygenase and the Arabidopsis dmr6-mutant was resistant to the DM pathogen Hyaloperonospora arabidopsidis (van Damme et al. 2008). In both tomato and cucumber, DMR6 orthologs have been identified, which could partially restore the DM resistance in the Arabidopsis dmr6-mutant (Zeilmaker (2012), indicating that tomato and cucumber have DMR6-like genes that provide susceptibility to DM.
PM and DM limit the production of cucumbers throughout the world (Morishita et al. 2003). Both dominant resistances and recessively inherited resistances to PM have been found in cucumber (Sitterly 1972; Morishita et al. 2003; He et al. 2013), whereas resistance to DM is in most of the cases inherited recessively in this crop (Olczak-Woltman et al. 2011). The recessively inherited resistances potentially resulted from loss-of-function of S-genes.
We describe here the search for MLO-, PMR- and DMR-like genes in cucumber that could be responsible for recessively inherited resistance. Zhou et al. (2013) searched for MLO-like genes in a previous version (version 1) of the published sequence of the cucumber genome. In this study, we used the improved version (version 2) of the cucumber genome to identify MLO-, PMR- and DMR-like genes in this crop. Further, we related the genetic loci of the putative S-genes with QTL for recessively inherited resistance to PM and/or DM in cucumber, described in literature.
Materials and methods
Identification of putative MLO-like proteins in cucumber
The amino acid sequences of the 15 MLO family members in the genome of A. thaliana (AtMLO1 to AtMLO15) were extracted from the NCBI protein database and used for blast searches for MLO-like predicted proteins of cucumber in the Cucurbit Genomics Database version 2 (www.icugi.org using the default blast search settings; Huang et al. 2009). For each MLO protein from A. thaliana, the best five hits in the cucumber database were stored in a list, and duplications were removed. The genomic positions of the yielded MLO-like genes in cucumber were extracted from the mentioned Cucurbit Genomics Database, using the genome browser.
Cluster analysis
Twelve orthologous MLO-proteins that have a proven function in susceptibility to powdery mildews were selected, i.e. HvMLO in barley (GenBank identification number P93766), OsMLO3 in rice (AAK94907), TaMLO_A1 and TaMLO_B1 in wheat (AAK94904 and AAK94905), AtMLO2, AtMLO6 and AtMLO12 in Arabidopsis (NP172598, NP176350, NP565902), SlMLO1 in tomato (NP001234814), PsMLO1 in pea (ACO07297), CaMLO2 in pepper (AFH68055), LjMLO1 in lotus (AAX77015), and MtMLO1 in barrel clover (ADV40949). For clade annotation according to Devoto et al. (2003), the sequences of the remaining MLO proteins in A. thaliana that are not known to be susceptibility proteins were added. All these sequences were aligned with the MLO-like putative proteins in cucumber, using the default settings in the CLC Main Workbench 6.8.4 (http://clcbio.com), and a phylogenetic tree was created using UGPMA clustering.
Conserved domains of Clade V proteins
Clade V contains all functionally proven MLO-like susceptibility proteins of other dicots (Fig. 1). The amino acid sequences of the cucumber proteins that clustered also in this clade were aligned to the sequences of these known susceptibility proteins of other dicots, using CLC. We regarded an amino acid as conserved if at least 7 out of 8 MLO-like susceptibility proteins of Clade V shared this amino acid, or if it was substituted with an amino acid with similar chemical properties according to the Rasmol colour scheme (Sayle 1994). We counted the number of these conserved amino acids in the susceptibility proteins, and counted for these amino acids the number of deviating amino acids in the Clade V proteins of cucumber. Further we counted for each known susceptibility gene the number of amino acids that deviated from the conserved amino acids in the other susceptibility genes. This allowed us to judge whether the degree of similarity of the cucumber Clade V proteins was within the variation range of the Clade V susceptibility genes for the conserved regions.
Positions of MLO-, PMR-, and DMR-like genes on the chromosomes of cucumber according to the cucumber genome browser version 2 in the Cucurbit Genomics Database. For the numbering of the genes we used the order of these genes on the chromosomes. The MLO-like genes that clustered in Clade V, and the PMR-like genes that showed highest homologies to PMR susceptibility genes in A. thaliana are underlined. QTL for PM or DM resistance are displayed as bars. Only QTL that explained more than 20 % of the variation are shown. The QTL names given by the authors are maintained (Sakata et al. 2006; Liu et al. 2008; Zhang et al. 2011, 2013; Fukino et al. 2013; He et al. 2013; Pang et al. 2013; Yoshioka et al. 2014). The main QTL for PM resistance detected by Sakata et al. (2006) is located on Chr. 1 according to the used SSR primer sequences, but in their paper this chromosome is named LG II
Transcriptomics
The ‘Chinese long’ inbred line 9,930 that was used for sequencing the whole cucumber genome (Huang et al. 2009), was cultivated in a greenhouse at Nickerson-Zwaan, The Netherlands. The plants were inoculated with the PM fungus P. fusca or the DM oomycete Pseudoperonospora cubensis. Leaf samples were harvested before inoculation, as well as 8, 24, 48, and 72 h after inoculation. Leaves were immediately frozen in liquid N2, ground. Material was sent to KeyGene, The Netherlands, for RNA-Seq. Total RNA was isolated using the Qiagen RNeasy Plant Mini Kit, following the manual included in the kit. Subsequently, RNA Seq libraries were made following the TruSeq™ RNA Sample Preparation v2 Guide protocol. After concentration measurement by qPCR (LightCycler® 480; Roche), the libraries were pooled, and sequenced using two lanes of the Illumina HiSeq 2000 sequencer. PhiX (~0.6 %) was spiked in according to manufacturer’s recommendations. The resulting reads were sorted into single fasta files per sample based on the sample tag sequences. The obtained read length was approximately 100 nt at a minimal read length of 36 nt. Per sample, 9.2–14.9 million reads were obtained. The transcript abundance per sample was assessed on the basis of the number of sequenced cDNA fragments per million of total reads sequenced (Reads per kilo base per million, RPKM; Kharchenko et al. 2011).
Identification of PMR-like and DMR-like proteins in cucumber
A. thaliana contains several non-MLO genes for susceptibility to PM and DM, so called PMR and DMR genes. The amino acid sequences of the five susceptibility genes PMR4 (Nishimura et al. 2003), PMR5 (Vogel et al. 2004), PMR6 (Vogel et al. 2002), DMR1 (van Damme et al. 2009), and DMR6 (van Damme et al. 2008) were obtained from the NCBI protein database, and used for blast searches in the Cucurbit Genomics Database version 2, using the default settings. Homologues from cucumber were included if score (bits) >1,000, or E-value < e−100. The genomic positions were extracted from the genome browser. Amino acid sequences of known DMR and PMR susceptibility genes were aligned to the homologous putative proteins in cucumber, using the default settings in the CLC Main Workbench 6.8.4 (http://clcbio.com), and phylogenetic trees were created using UGPMA clustering. Also the functionally analysed susceptibility genes SlPMR4 and SlDMR1 discovered in tomato (Huibers et al. 2013) were included in the phylogenetic trees.
Results
Cucumber has 13 members in the MLO family
The search for putative MLO-like proteins in the cucumber genome, using the 15 A. thaliana MLO proteins, yielded 13 putative members of the MLO family (Table 1). All chromosomes appear to contain at least one MLO-like gene, apart from chromosome 7, which lacks such genes (Fig. 1). Most MLO-like genes are scattered among the genome, but in three cases two genes are located close together, i.e. the pairs CsaMLO1 and CsaMLO2, CsaMLO4 and CsaMLO5, and CsaMLO8 and CsaMLO9 (Fig. 1). For these three cases of tight linkage, the genes were not tandem repeats, but clustered in different clades (Fig. 2).
Phylogenetic tree of 13 MLO-like putative proteins in cucumber, 12 MLO-like proteins known to be required for susceptibility to powdery mildew (PM) in monocots or dicots, and 12 MLO-like proteins in A. thaliana that are not known as susceptibility proteins. The numbering of the six clades is according to Devoto et al. (2003). The proteins that have been reported as required for susceptibility to PM in monocotyledons or dicotyledons are highlighted. Bootstrap values at 100 replicates are displayed
CsaMLO1, CsaMLO8 and CsaMLO11 cluster with the known MLO-like susceptibility proteins in dicots
The amino acid sequences of the 13 MLO-like putative proteins in cucumber were aligned to the sequences of the 15 MLO proteins in A. thaliana, and the 12 MLO proteins that have a proven function in susceptibility to PM in monocots or dicots. This multiple alignment was used for construction of the phylogenetic tree shown in Fig. 2. The six clades were numbered according to the annotation by Devoto et al. (2003). All MLO proteins that have proven to play a role in susceptibility to PM in monocots, i.e. in barley, rice and wheat, group in Clade IV, and all MLO proteins that are important for susceptibility to PM in dicots group in Clade V (Fig. 2), which resembles the results shown by others, such as Devoto et al. (2003) and Acevedo-Garcia et al. (2014). Three putative proteins (CsaMLO1, CsaMLO8, and CsaMLO11) from cucumber, which is a dicot, were positioned in this Clade V (Fig. 2). Based on this phylogenetic tree, the three underlying genes CsaMLO1, CsaMLO8 and CsaMLO11 are considered as candidate genes for susceptibility to PM.
All three candidates harbour the conserved regions of MLO-like susceptibility proteins
We aligned the amino acid sequences of the three predicted cucumber proteins from Clade V (CsaMLO1, CsaMLO8, and CsaMLO11) to the sequences of the known MLO-like susceptibility proteins from the same clade (Fig. 2, and Online Resource 1). The three Clade V cucumber proteins showed 97.3–98.5 % similarity to the susceptibility proteins in other dicots in the conserved regions (Table 2). This similarity of the cucumber proteins is well within the range of conservation of the proven susceptibility proteins of dicots (94.2–98.9 %), as far as the conserved regions are concerned. The LjMLO1 protein from lotus deviated more (94.2 %) in these regions, but in spite of that is still a functional susceptibility gene, according to Humphry et al. (2011). Further, the multiple alignment illustrates that none of the three cucumber genes harbours an early stop codon that would have given rise to a truncated protein. Therefore, none of the three cucumber proteins can be excluded as a candidate gene based on absence of one or more conserved regions.
CsaMLO1 expression is upregulated after PM inoculation
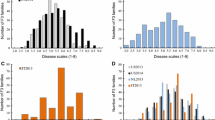
A characteristic of MLO-susceptibility genes is that their transcript abundances are increased about 8 h after inoculation with the PM causing fungus (e.g. Zheng et al. 2013). Therefore we investigated the expression of the MLO-like genes in cucumber after inoculation with PM, using RNA-Seq. Only the expression of CsaMLO1 was clearly upregulated 8 h after inoculation (Fig. 3).
Transcript abundances of MLO- and DMR6-like genes in cucumber leaves after inoculation with the pathogen, measured by means of RNA-Seq. a Transcript abundances of MLO-like genes upon powdery mildew (PM) infection. b Transcript abundances of DMR6-like genes upon downy mildew (DM) infection. The transcript abundances before inoculation were set at 1
PMR- and DMR-like genes in the cucumber genome
We searched for homologs of the PMR4, PMR5 and PMR6 susceptibility genes for PM. These genes do not belong to the MLO-gene family. For PMR4 in A. thaliana we found ten homologous predicted proteins in cucumber (Table 3), named CsaPMR4-1 to 10. Two predicted proteins (CsaPMR4-2 and CsaPMR4-9) appeared to be highly similar to AtPMR4 (Fig. 4). For PMR5, only one homologous protein was found in cucumber, which we named CsaPMR5 (Table 3). For PMR6, 13 homologous predicted proteins were detected. Construction of a phylogenetic tree for these 13 predicted proteins revealed that CsaPMR6-5, 12 and 13 show the highest homologies to the PMR6 susceptibility protein in A. thaliana. For DMR1 and DMR6 we found only one and two homologous proteins, respectively (Table 3).
Phylogenetic trees of PMR-like proteins in cucumber. a Ten PMR4-like putative proteins in cucumber, and the PMR4 protein, required for susceptibility to powdery mildew in A. thaliana; b 13 PMR6-like putative proteins in cucumber, and the PMR6 susceptibility protein in A. thaliana. Bootstrap values at 100 replicates are displayed
We consulted literature on induction of expression of PMR4, PMR5, PMR6, DMR1, DMR6 by PM or DM, and did not find indications of elevated transcript abundance after infection, apart from DMR6 in Arabidopsis. This gene was locally induced after infection with DM in A. thaliana (van Damme et al. 2008). Therefore we analyzed the expression to DMR6 upon infection. However, the CsaDMR6-like genes did not show a strong induction (Fig. 3b).
Discussion
The cucumber MLO-like genes as potential S-genes to PM
We searched for MLO-like genes in the cucumber genome, and found 13 putative MLO-like genes, scattered among the chromosomes (Table 1; Fig. 1). Out of these 13 MLO-like genes, three (CsaMLO1, CsaMLO8, and CsaMLO11) code for predicted proteins that belong to Clade V (Fig. 2). Not all MLO-like genes that cluster in this clade are susceptibility genes (Zheng et al. 2013), but until now all MLO-like genes of dicots that have shown to be required for susceptibility to PM, belong to Clade V (Fig. 2). Therefore, the clustering in this clade is a strong selection criterion for MLO-like susceptibility genes. The predicted proteins from these three genes appear to have high similarity to proven MLO-like susceptibility genes, and none of the three candidate could be excluded based on deviation in conserved regions or an early truncation of the protein (Online Resource 1).
Transcript abundances of MLO-like susceptibility genes tend to increase after PM infection already 5 h after inoculation (e.g. Piffanelli et al. 2002; Zheng et al. 2013). Therefore we investigated the transcript abundances of all MLO-like genes in cucumber, using RNA-Seq. Only the expression of CsaMLO1 was upregulated soon after PM infection. CsaMLO1 is one of the three putative genes for which the predicted proteins belong to Clade V. This implies that CsaMLO1 is the most likely candidate for being a susceptibility gene for PM in cucumber according to the transcript induction. However, Zheng et al. (2013) also found an MLO-like gene in pepper that was not induced by PM, but still seemed to be a susceptibility gene. Therefore, induction of expression should not be regarded as an absolute selection criterion.
In a patent application Diergaarde et al. (2012) describe a functional test of three Clade V MLO-genes from melon (Cucumismelo). They used these genes for complementation of mlo mutants of A. thaliana, lacking two out of three functional MLO susceptibility genes (Atmlo2 and Atmlo6) or lacking all three functional genes (Atmlo2, 6 and 12). The complementation by one of these genes, named CmMLO1, partially restored the PM susceptibility in A. thaliana, indicating that CmMLO1 is a S-gene (Diergaarde et al. 2012). Cucumber and melon are closely related and belong to the same genus. We aligned the putative cDNA sequence of CmMLO1 with the putative cDNA sequences of the three Clade V genes of cucumber. The phylogenetic tree revealed that CmMLO1 is most related to CsaMLO8 (Online Resource 2). This result provides evidence that CsaMLO8 is also a likely candidate.
Zhou et al. (2013) also searched for MLO-like gene in the cucumber genome. They used an older version (version 1) of the published genome sequence of cucumber ‘9930’, compared to the sequence we used (version 2). As the gene ID codes changed between these versions, direct comparison is a bit hampered. Zhou et al. detected 14 MLO-like sequences, whereas we detected one less. They indicated that Chr. 1 contains three MLO-like genes, whereas we detected here only two MLO-candidates (Fig. 1). The predicted ORFs of two neighbouring MLO-like genes on this chromosome mentioned by Zhou et al. are shorter (513 and 1,080 bp) than the ORFs of the other predicted MLO-like genes (1,606 bp on average), and the number of transmembrane domains was only one and three respectively, although MLO-proteins usually have about seven transmembrane domains (Devoto et al. 2003). However, version 2 of the genome browser combines the two short genes into one gene (i.e. Csa1M086900.1 = CsaMLO2), with a ORF size of 1,755 bp that is within the normal range of MLO-like genes. Zhou et al. indicated that the two short MLO-like genes coded for proteins that clustered in Clade VI and a new Clade VII. However, the CsaMLO2 predicted in the version 2 cucumber genome clusters in Clade VI (Fig. 2). Therefore, the new Clade VII of Zhou et al., which was also mentioned by Acevedo-Garcia et al. (2014), probably resulted from an error in version 1 of the genome sequence, and refers to a fragment of a Clade VI protein.
In view of the references to the older genome sequence, the two short genes on Chr 1 that should be one gene, and some lengthy gene (such as CsMLO1415) coded by Zhou et al., we decided to keep our own numbering for MLO-like genes. Also we decided to use Csa as genus code rather than Cs, in view of consistency with the genus code Csa applied for all putative genes in the whole genome sequence (Huang et al. 2009).
Co-localization of MLO-, PMR-like genes with QTLs for resistance to PM in cucumber
In Fig. 1 we depicted QTL for PM resistance, detected by others. We limited to QTL that explained more than 20 % of the variation. The QTL found by Sakata et al. (2006) and He et al. (2013) represent recessively inherited resistances, but for the other QTL it is not clear from the papers whether the resistances inherited dominantly or recessively. The CsaMLO1 gene is located at the border of the PM resistance region detected by Fukino et al. (2013), and might be the causal gene for these QTL. However, a more detailed mapping of the QTL or a functional analysis of CsaMLO1 would be required to provide an answer on the question whether a loss of function mutation in CsaMLO1 caused this QTL, or whether such a mutation in this gene or in its regulatory sequences may cause resistance in other genotypes.
At the position of CsaMLO8, a second Clade V gene, He et al. (2013) detected a strong, recessively inherited QTL for PM resistance from the cucumber-inbred line WI 2757. CsaMLO8 is an interesting candidate gene for this QTL. This is also supported by the functional analysis of the CmMLO1 gene in melon by Diergaarde et al. (2012), as discussed above.
Liu et al. (2008) found a QTL for PM resistance at CsaMLO13, but this gene does not cluster in Clade V (Fig. 2), nor was it induced by PM infection (Fig. 3), and consequently it is a less likely candidate gene for the PM QTL. On Chr. 5 three other QTL for PM resistance are displayed, close to CsaPMR6-10 (Fig. 1). However, this candidate gene is less homologous to AtPMR6 compared to some other CsaPMR6-homologs (Fig. 4b).
PMR4, PMR5, and PMR6 are required for susceptibility to PM in A. thaliana and tomato (Nishimura et al. 2003; Vogel et al. 2002, 2004; Huibers et al. 2013).We found a series of PMR4-, PMR5-, and PMR6-like genes in the cucumber genome. Six out of these showed high homology to the known PMR-genes. Two out of these six genes (CsaPMR4-2 and CsaPMR5) are located at the borders of QTL on chromosome 1 (Fig. 1), and one (CsaPMR6-12) is located in the strong recessively inherited QTL for PM resistance at the bottom of chromosome 5, detected by He et al. (2013). The last PMR6-like gene is closely linked to the Clade V gene CsaMLO8, and both are serious candidates that could explain the strong QTL detected. For the mentioned QTL, a more thorough analysis is required for investigation of a hypothetical causal relationship between a possible loss of function of one of the mentioned candidate susceptibility genes at these loci and the recessively inherited QTL.
Co-localization of DMR-like genes with QTLs for resistance to DM in cucumber
The number of DMR-like genes is far lower compared to PMR-like genes, as only one DMR1-like gene was detected in the cucumber genome, and only two DMR6-like genes (Table 3; Fig. 1). DMR1 and DMR6 are susceptibility genes for DM in A. thaliana (van Damme et al. 2008, 2009). A homologous gene of DMR1 in tomato (SlDMR1) appeared to be required for PM susceptibility in that crop (Huibers et al. 2013). Several QTL for DM in cucumber have been described, listed in a review paper from Olczak-Woltman et al. (2011). The majority of the DM resistances in cucumber are inherited recessively (Olczak-Woltman et al. 2011), which indicates the presence of susceptibility genes. We selected for QTL that explained more than 20 % of the variation, and could be positioned on the genome map, using primer sequences and the cucumber genome sequence. We found a QTL that met these criteria at the top of chromosome 5 (Fig. 1; Pang et al. 2013). A recently described QTL with recessively inheriting resistance to DM was detected at the same location by Yoshioka et al. (2014). Surprisingly, also CsaDMR6-2 resides at this location. Also for this gene, it would be worthwhile to study whether a loss of function mutation, or a low expression level of this gene caused the resistance.
Zeilmaker (2012) cloned a DMR6-like gene from cucumber, and over-expressed it under control of the CaMV 35S promoter in a dmr6 mutant of A. thaliana. This restored to some extent the susceptibility to DM, indicating that the cloned DMR6-like gene from cucumber is indeed a susceptibility gene to DM. We aligned the sequences of the primers that Zeilmaker used to amplify this cucumber gene, and found that he had amplified the DMR6-like gene on Chr 4 (Csa4M091870.1), that we named CsaDMR6-1 (Table 3). Surprisingly, at the position of CsaDMR6-1 no strong QTL for DM resistance was mapped yet (Fig. 1).
Zhang et al. (2013) also mapped DM resistance in cucumber, and found several weak QTL in three consecutive years. The QTL on Chr 5 appeared in all 3 years. Therefore we included it in Fig. 1, although it explained slightly less than 20 % of the variation. However, the QTL interval is besides the CsaDMR6-2 gene (Fig. 1), and therefore does not support this gene as the candidate gene in the resistant parent used by Zhang et al. (2013).
We summarize that analysis of the cucumber genome yielded several candidate genes for susceptibility to PM, i.e. CsaMLO1, CsaPMR4-2, and CsaPMR5 on chromosome 1, and CsaMLO8 and CsaPMR6-12 at the bottom of chromosome 5. These candidate genes are located at QTL for recessively inherited resistance to PM. Two strong QTL for DM resistance at the top of chromosome 5 may be caused by CsaDMR6-2, in view of their genetic locations, and in view of the recessive inheritance of the resistances.
References
Acevedo-Garcia J, Kusch S, Panstruga R (2014) Magical mystery tour: MLO proteins in plant immunity and beyond. New Phytol. doi:10.1111/nph.12889
Bai Y, Pavan S, Zheng Z et al (2008) Naturally occurring broad-spectrum powdery mildew resistance in a Central American tomato accession is caused by loss of MLO function. Mol Plant Microbe Interact 21:30–39. doi:10.1094/MPMI-21-1-0030
Cohen R, Burger Y, Katzir N (2004) Monitoring physiological races of Podosphaera xanthii (syn. Sphaerotheca fuliginea), the causal agent of powdery mildew in cucurbits: Factors affecting race identification and the importance for research and commerce. Phytoparasitica 32:174–183. doi:10.1007/BF02979784
Consonni C, Humphry ME, Hartmann HA et al (2006) Conserved requirement for a plant host cell protein in powdery mildew pathogenesis. Nat Genet 38:716–720. doi:10.1038/ng1806
Devoto A, Hartmann HA, Piffanelli P et al (2003) Molecular phylogeny and evolution of the plant-specific seven-transmembrane MLO family. J Mol Evol 56:77–88. doi:10.1007/s00239-002-2382-5
Diergaarde PJ, van Enckevort LJG, Posthuma KI, Prins MW (2012) Powdery mildew resistance providing genes in Cucumismelo. Patent appl. WO 2012/116938
Ellinger D, Naumann M, Falter C et al (2013) Elevated early callose deposition results in complete penetration resistance to powdery mildew in Arabidopsis. Plant Physiol 161:1433–1444. doi:10.1104/pp.113.900456
Fukino N, Yoshioka Y, Sugiyama M et al (2013) Identification and validation of powdery mildew (Podosphaeraxanthii)-resistant loci in recombinant inbred lines of cucumber (Cucumissativus L.). Mol Breed 32:267–277. doi:10.1007/s11032-013-9867-3
Fukuoka S, Saka N, Koga H et al (2009) Loss of function of a proline-containing protein confers durable disease resistance in rice. Science 325:998–1001. doi:10.1126/science.1175550
He X, Li Y, Pandey S et al (2013) QTL mapping of powdery mildew resistance in WI 2757 cucumber (Cucumis sativus L.). Theor Appl Genet 126:2149–2161. doi:10.1007/s00122-013-2125-6
Huang S, Li R, Zhang Z et al (2009) The genome of the cucumber, Cucumissativus L. Nat Genet 41:1275–1281. doi:10.1038/ng.475
Huibers RP, Loonen AEHM, Gao D et al (2013) Powdery mildew resistance in tomato by impairment of SlPMR4 and SlDMR1. PLoS One 8:e67467. doi:10.1371/journal.pone.0067467.s001
Humphry M, Reinstädler A, Ivanov S et al (2011) Durable broad-spectrum powdery mildew resistance in pea er1 plants is conferred by natural loss-of-function mutations in PsMLO1. Mol Plant Pathol 12:866–878. doi:10.1111/j.1364-3703.2011.00718.x
Kharchenko PV, Xi R, Park PJ (2011) Evidence for dosage compensation between the X chromosome and autosomes in mammals. Nat Genet 43:1167–1169. doi:10.1038/ng.991
Kim DS, Hwang BK (2012) The pepper MLO gene, CaMLO2, is involved in the susceptibility cell-death response and bacterial and oomycete proliferation. Plant J 72:843–855. doi:10.1111/tpj.12003
Kim MC, Panstruga R, Elliott C et al (2002) Calmodulin interacts with MLO protein to regulate defence against mildew in barley. Nature 416:447–451. doi:10.1038/416447a
Liu L, Yuan X, Cai R et al (2008) Quantitative trait loci for resistance to powdery mildew in cucumber under seedling spray inoculation and leaf disc infection. J Phytopathol 156:691–697. doi:10.1111/j.1439-0434.2008.01427.x
Lyngkjær MF, Newton AC, Atzema JL, Baker SJ (2000) The Barley mlo-gene: an important powdery mildew resistance source. Agronomie 20:745–756. doi:10.1051/agro:2000173
Morishita M, Sugiyama K, Saito T, Sakata Y (2003) Powdery mildew resistance in cucumber. JARQ 37(1):7–14
Nishimura MT, Stein M, Hou BH, Vogel JP, Edwards H (2003) Loss of a callose synthase results in salicylic acid-dependent disease resistance. Science 301:969–972. doi:10.1126/science.1086716
Olczak-Woltman H, Marcinkowska J, Niemirowicz-Szczytt K (2011) The genetic basis of resistance to downy mildew in Cucumis spp.—latest developments and prospects. J Appl Genet 52:249–255. doi:10.1007/s13353-011-0030-8
Pang X, Zhou X, Wan H, Chen J (2013) QTL mapping of downy mildew resistance in an introgression line derived from interspecific hybridization between cucumber and Cucumis hystrix. J Phytopathol 161:536–543. doi:10.1111/jph.12103
Pavan S, Jacobsen E, Visser RGF, Bai Y (2009) Loss of susceptibility as a novel breeding strategy for durable and broad-spectrum resistance. Mol Breed 25:1–12. doi:10.1007/s11032-009-9323-6
Pavan S, Schiavulli A, Appiano M et al (2011) Pea powdery mildew er1 resistance is associated to loss-of-function mutations at a MLO homologous locus. Theor Appl Genet 123:1425–1431. doi:10.1007/s00122-011-1677-6
Piffanelli P, Zhou F, Casais C et al (2002) The barley MLO modulator of defense and cell death is responsive to biotic and abiotic stress stimuli. Plant Physiol 129:1076–1085
Sakata Y, Kubo N, Morishita M et al (2006) QTL analysis of powdery mildew resistance in cucumber (Cucumis sativus L.). Theor Appl Genet 112:243–250. doi:10.1007/s00122-005-0121-1
Santo T, Rashkova M, Alabaça C, Leitão J (2013) The ENU-induced powdery mildew resistant mutant pea (Pisumsativum L.) lines S(er1mut1) and F(er1mut2) harbour early stop codons in the PsMLO1 gene. Mol Breed 32:723–727. doi:10.1007/s11032-013-9889-x
Sayle R (1994) RasMol v 2.6 manual. http://life.nthu.edu.tw/~fmhsu/rasframe/COLORS.HTM#aminoc
Sitterly WR (1972) Breeding for disease resistance in cucurbits. Annu Rev Phytopathol 10:471–490. doi:10.1146/annurev.py.10.090172.002351
Torés JA, Sánchez-Pulido JM, López-Ruiz F et al (2009) First occurrence of cucurbit powdery mildew caused by race 3-5 of Podosphaera fusca in Spain. Plant Dis 93:1073. doi:10.1094/PDIS-93-10-1073B
van Damme M, Huibers RP, Elberse J, van den Ackerveken G (2008) Arabidopsis DMR6 encodes a putative 2OG-Fe(II) oxygenase that is defense-associated but required for susceptibility to downy mildew. Plant J 54:785–793. doi:10.1111/j.1365-313X.2008.03427.x
van Damme M, Zeilmaker T, Elberse J et al (2009) Downy mildew resistance in Arabidopsis by mutation of HOMOSERINE KINASE. Plant Cell 21:2179–2189
Várallyay É, Giczey G, Burgyán J (2012) Virus-induced gene silencing of Mlo genes induces powdery mildew resistance in Triticum aestivum. Arch Virol 157:1345–1350. doi:10.1007/s00705-012-1286-y
Vogel JP, Raab TK, Schiff C, Somerville SC (2002) PMR6, a pectatelyase–like gene required for powdery mildew susceptibility in Arabidopsis. Plant Cell 14:2095–2106
Vogel JP, Raab TK, Somerville CR, Somerville SC (2004) Mutations in PMR5 result in powdery mildew resistance and altered cell wall composition. Plant J 40:968–978. doi:10.1111/j.1365-313X.2004.02264.x
Yoshioka Y, Sakata Y, Sugiyama M, Fukino N (2014) Identification of quantitative trait loci for downy mildew resistance in cucumber (Cucumissativus L.). Euphytica. doi:10.1007/s10681-014-1102-8
Zeilmaker T (2012) Functional and applied aspects of the DOWNY MILDEW RESISTANT 1 and 6 genes in Arabidopsis. PhD Thesis, Univ. of Utrecht, The Netherlands, pp 1–147
Zhang S, Liu M, Miao H et al (2011) QTL mapping of resistance genes to powdery mildew in cucumber (Cucumis sativus L.). Scientia Agri Sinica 44:3584–3593
Zhang SP, Liu MM, Miao H et al (2013) Chromosomal mapping and QTL analysis of resistance to downy mildew in Cucumis sativus. Plant Dis 97:245–251
Zheng Z, Nonomura T, Appiano M et al (2013) Loss of function in Mlo orthologs reduces susceptibility of pepper and tomato to powdery mildew disease caused by Leveillula taurica. PLoS One 8:e70723. doi:10.1371/journal.pone.0070723
Zhou SJ, Jing Z, Shi JL (2013) Genome-wide identification, characterization, and expression analysis of the MLO gene family in Cucumissativus. Genet Mol Res 12:6565–6578. doi:10.4238/2013
Acknowledgments
This work was funded in part by the Technological Top Institute Green Genetics, the Netherlands (TTI-GG project INTCFD040RP), together with Nickerson-Zwaan, Nunhems Netherlands, Takii Europe B.V., and Rijk Zwaan B.V. Dr Jan-Peter Nap (Applied Bioinformatics, Plant Research International, Wageningen University and Research Centre) provided valuable comments to the manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Schouten, H.J., Krauskopf, J., Visser, R.G.F. et al. Identification of candidate genes required for susceptibility to powdery or downy mildew in cucumber. Euphytica 200, 475–486 (2014). https://doi.org/10.1007/s10681-014-1216-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-014-1216-z