Abstract
Interspecific hybridization of various tuberous Begonia species hybrids with Begonia socotrana results in so-called ‘Elatior’-begonias hybrids (B. × hiemalis Fotsch). In our study, genomic in situ hybridization (GISH) has been employed to assess the genome composition in eleven ‘Elatior’-begonias hybrids and their ancestor genotypes. Genomic DNA of tuberous Begonia was sonicated to 1–10-kb fragments, labelled by nick translation with digoxigenin-11-dUTP and used as a probe whereas B. socotrana DNA was autoclaved to 100 bp fragments and used as block. The genome of tuberous Begonia was clearly pronounced in ‘Elatior’-begonias when the probe concentration was ~3.75 ng/μl (150 ng/slide), with 30 times the excess of B. socotrana blocking DNA and stringency of post hybridization washings at 73% (0.1× SSC at 42°C). In ‘Elatior’-begonias hybrids GISH distinguished two groups comprising short (0.6–1.03 μm in length) and relatively longer chromosomes (1.87–3.88 μm) which represent B. socotrana and tuberous Begonia genomes, respectively. The number of chromosomes derived from tuberous Begonia ranged from 14 to 56 and for B. socotrana from 7 to 28 which suggest the presence of different ploidy levels in analyzed ‘Elatior’-begonia hybrids. Intergenomic recombination has not been detected through GISH in hybrids analyzed. Genomic in situ hybridization turned out to be useful to identify the genome constitution of ‘Elatior’-begonia hybrids and thus gain an insight into the origins of these cultivars. This knowledge on the ploidy level and genome composition is essential for further progress in breeding Begonias.
Similar content being viewed by others
Introduction
The genus Begonia, a member of the family Begoniaceae, includes around 1500 species distributed mainly in tropical and subtropical regions (Doorenbos et al. 1998; Wagner 1999). Begonia species are widely hybridized for improving flowering and foliage of plants where interspecific hybridization is the most suitable method for the transmission into new cultivars of important agricultural traits, such as winter flowering capacity, new flower shapes and colours and disease resistance. The current commercial assortment comprise mostly of ‘Elatior-hybrids’ (B. × hiemalis Fotsch) (over 100 varieties) representing around 88% of the total Begania production (Kroon 1993). The early varieties of ‘Elatior’-begonias resulted from crosses between winter flowering Begonia socotrana Hook and ‘Viscoutess Doneraile’, a very early hybrid tuberous Begonias (B. × tuberhybrida Voss) (Arends 1970; Doorenbos 1973). However, the majority of modern varieties of ‘Elatior’-begonias are the result of crossing various tuberous Begonia species hybrids (B. × tuberhybrida Voss) and B. socotrana Hook. F (Gleed 1961). Some varieties of ‘Elatior-hybrids’ have also been obtained from the following crosses: ‘Elatior’-begonia × B. socotrana and ‘Elatior’-begonia × tuberhybrid (Arends 1970).
Chromosome numbers have been reported for many species of Begonia, ranging from 2n = 18, 22, 26, 28, 30, 38, 41, 44, 46, 52 to 82 (Okuno and Nagai 1954; Zeilinga 1962; Ye et al. 2004; Nakata et al. 2007) which suggests the high levels of polyploidy and aneuploidy in the genus. The progenitors of ‘Elatior’-begonia hybrids, the tuberous cultivars have originated from hybrids between a number of widely diverging wild species with 2n = 26 and 2n = 28 chromosomes (Legro and Doorenbos 1969). By breeding and selection B. × tuberhybrida has been brought to polyploid level (Kroon 1993). In the hybrid tuberous Begonias (B. × tuberhybrida Voss) several different chromosome numbers were found (2n from 27 to 64) (Okuno and Nagai 1954; Legro and Haegeman 1971). B. socotrana, the second ancestral species of ‘Elatior’-begonia hybrids, comprises of 2n = 2× = 28 chromosomes (Matsuura and Okuno 1936; Doorenbos and Legro 1968).
Chromosomes of begonia are relatively small ranging in length from 0.5 μm in B. socotrana (Arends 1970) to 5.43 μm in B. coptidifolia (Ye et al. 2004) and poorly differentiated (Legro and Haegeman 1971). Due to small size and similar morphology, the evaluation of chromosome number and the discrimination of genomes in hybrids can be liable to mistakes (Legro and Doorenbos 1969). For some Begonia genotypes different chromosome numbers have been established by different authors (Zeilinga 1962). For this reason verification of Begonia hybrids based on chromosome number or karyomorphology may be problematic.
Genomic in situ hybridization (GISH), which uses total genomic DNA of one of the parents as the probe, is a very effective molecular cytogenetic method usually providing clear and unambiguous distinction between genomes. This technique is especially effective in plants with large chromosomes such as in Lilium (Barba-Gonzalez et al. 2006), Alstroemeria (Kamstra et al. 1999) or Tulipa (Marasek et al. 2006). However, it has been also successfully applied to differentiate genomes with small chromosomes such as in Oryza (Li et al. 2001), Lycopersicon (Haider Ali et al. 2002), Brassica (Hasterok et al. 2005), and Arabidopsis (Ali et al. 2004). As yet, genomic in situ hybridization has not been applied to analyze hybrids of the genus Begonia.
The aim of present study was to optimize GISH technique for Begonia chromosomes and to identify B. socotrana and Tuberous hybrids genomes in ‘Elatior’-begonia hybrids of different ploidy level thus gaining an insight into the origins of these cultivars as well as the basic chromosome number.
Materials and methods
Plant materials
Begonia socotrana, two tuberous Begonia hybrids and eleven different accessions of ‘Elatior’-begonias hybrids were used in this study for GISH analysis (Table 2). All genotypes were provided by Beekenkamp B.V., Maasdijk, the Netherlands. For analysis of chromosome morphology and adaptation of GISH technique for Begonia chromosomes, tetraploid tuberous Begonia characterized by erect plant, totally filled red flowers and tubers has been selected as a representative genotype of tuberous Begonias. The cuttings from all hybrids and species were rooted in greenhouse under standard growing condition applicable for begonia cultivation (20–25°C) and being maintained in Wageningen UR, Plant Breeding, The Netherlands.
Chromosome preparation
Root tips were pre-treated with 0.05% colchicine for 2.5 h and then fixed in 3:1 ethanol:glacial acetic acid. The roots were subjected to enzymatic digestion in a mixture comprising 0.2% (w/v) pectolyase Y23, 0.2% (w/v) cellulase RS at 37°C for about 2 h. Meristems were squashed in a drop of 45% acetic acid. After freezing in liquid nitrogen, cover slips were removed by using a razor blade and the preparations were dehydrated in absolute ethanol and air dried. The best slides were selected under a phase contrast microscope (Leica Dialux 20 EB) and stored at −20°C until use.
Preparation of probe and block DNA
Total genomic DNA of B. socotrana and tuberous Begonia was extracted from young leaves using modified method of Fulton et al. (1995). The isolation was preceded by the initial wash step of powdered leave tissue with TE buffer (10 mM tris-HCL, 10 mM EDTH, pH 8) according to Kopperud and Einset (1995). Tuberous Begonia DNA were sonicated to 1–10-kb fragments and labeled by nick translation with digoxigenin-11-dUTP by a standard nick translation protocol (Roche Diagnostics GmbH, Mannheim, Germany). Block DNA of B. socotrana was obtained by autoclaving for 5 min to a fragments 100–300 bp.
Genomic in situ hybridization and detection
DNA denaturation and in situ hybridization steps were performed according to Hasterok et al. (2001) and Marasek et al. (2006). Slides were pre-treated with RNase A (100 μg/ml) for 1 h at 37°C, treated with 10 mM HCl at 37°C for 2 min followed by incubation in pepsin solution (5 μg/ml) for 10 min and post-fixed in 1% formaldehyde in PBS buffer for 10 min. The hybridization mixture consisted of 50% deionized formamide, 10% dextran sulphate, 2× SSC, 1% SDS, 150 ng of probe DNA per slide and block DNAs (B. socotrana DNA and herring sperm DNA) in 30- to 60-fold excess of labeled probe. The hybridization mix was denatured for 10 min at 75°C and placed on ice for 10 min. After the hybridization mix was add to the slides, a 4.5 min denaturation step at 70°C was carried out. Hybridization was done overnight in a humid chamber at 37°C. The post-hybridization washes were carried out for 15 min in 2× SSC at room temperature, followed by washes in 0.1× SSC at 42°C for 30 min (73% stringency) and 2× SSC for 15 min at room temperature. Digoxigenin-labelled DNA was detected with antidigoxigenin-FITC (sheep) (Boehringer, Mannheim, Germany) and amplified with anti-sheep-FITC (rabbit) (Vector Laboratories). The chromosomes were counterstained with 1 μg/ml 4,6-diamidino-2-phenylindole (DAPI, Sigma) in Vectashield (Vector Laboratories).
Image capturing and processing
Images of fluorescently stained chromosomes were acquired using a Canon digital camera attached to an Axiophot microscope with an appropriate filter and then processed using software (Axio Vision 4.2). For each genotype 8–15 metaphases have been analysed at different stages of chromosomes contraction. Chromosome length was determined using freeware application MicroMeasure available on the Internet at the http://www.colostate.edu/Depts/Biology/MicroMeasure.
Results
GISH technique in Begonia
In this study, we have attempted to isolate DNA from begonia leaves using several conventional procedures (Aljanabi and Martinez 1997; Murray and Thompson 1980; Fulton et al. 1995). None of these methods yielded above 5 μg DNA per gram fresh weight of begonia leaves both for B. socotrana and tuberous Begonia cultivar. The initial washing step of powdered leaf tissue in T10E10 buffer according to Kopperud and Einset (1995) increased the yield to 20–30 μg of DNA per gram fresh weight of leaves.
Initially, the total genomic DNA of both tuberous Begonia cultivar and B. socotrana labeled with digoxigenin-11-dUTP and biotin-11-dUTP respectively, were used as hybridization probes to differentiate the respective chromosomes in the cells of tuberous Begonia, B. socotrana and ‘Elatior’-begonias (Table 1). Since genomic probe of B. socotrana did not label chromosomes of B. socotrana, digoxigenin labeled genomic probe of tuberous Begonia were used exclusively for differentiation of the chromosomes in ‘Elatior’-begonias. The genome of tuberous Begonia was clearly pronounced in ‘Elatior’-begonias when the probe concentration was ~3.75 ng/μl (150 ng/slide), with 30 times the excess of B. socotrana blocking DNA and stringency of post hybridization washings at 73% (0.1× SSC at 42°C).
Chromosome morphology of the ancestral genotypes of ‘Elatior’-begonia hybrids
Figure 1 shows the somatic metaphase chromosomes of tuberous Begonia (2n = 4x = 56) and B. socotrana (2n = 2x = 28). Tuberous Begonia chromosomes were labeled uniformly when using total genomic DNA of tuberous Begonia as a probe (FITC-green fluorescence) (Fig. 1a). Chromosomes were small ranging from 1.87 to 3.88 μm and little differentiated. Centromeres were distinct only for some chromosomes when they were less condensed at prometaphase stage. Figure 1b represents the chromosome complement of B. socotrana stained with DAPI (blue fluorescence). The chromosomes at metaphase were particularly small ranging from 0.6 to 1.03 μm in length. The position of the centromeres was not discernible. Both for chromosomes of tuberous Begonia and B. socotrana it seems impossible to identify homologous pairs due to the lack of differentiation of chromosomes.
Chromosomes differentiation in ‘Elatior’-begonia hybrids as revealed by GISH
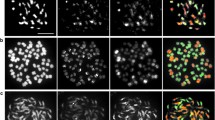
The genome constitution of analyzed genotypes based on the results of genomic in situ hybridization and their origin are summarized in Table 2. GISH clearly distinguished two parental genomes in all ‘Elatior’-begonia hybrids analysed (Fig. 2a–j) except for genotype 04-122-04 (2n = 44) in which chromosomes derived from tuberous Begonia were found exclusively (Fig. 2k). Figure 2l represents tuberous Begonia hybrid 04-117-04 (2n = 4x = 56) resulted from crosses between tetraploid forms of tuberous Begonia. As genomic DNA of tuberous Begonia was used as a probe in this way green fluorescence (FITC) represented the chromosomes of tuberous Begonia whereas short B. socotrana chromosomes were stained by DAPI (blue fluorescence). In ‘Elatior’-begonia hybrids, the number of chromosomes derived from tuberous Begonia ranged from 14 to 56 and from B. socotrana from 7 to 28. In three out of 12 genotypes analyzed (genotypes HO-7256-3, HO-7256-4, H08-124-01) GISH revealed that 14 chromosomes were inherited from tuberous Begonia and 14 chromosomes from B. socotrana (Fig. 2a–c). Four genotypes of ‘Elatior’-begonia (‘Netja Dark’, H07-102-13, H07-III-02, H08-143-03) had in total 2n = 42 chromosomes, 28 long chromosomes belonging to tuberous Begonia (green fluorescence) and 14 short chromosomes of B. socotrana (blue fluorescence) (Fig. 2d–g). In hybrid 04-187-23 (2n = 63) it was possible to distinguish 56 chromosomes of tuberous Begonia and 7 chromosomes of B. socotrana (Fig. 2h). In genotype H08-132-01, GISH showed 2n = 56 chromosomes of which 28 represented tuberous Begonia genome and 28 B. socotrana genome (Fig. 2i) whereas in H08-143-01 (2n = 46) 27 chromosomes were derived from tuberous Begonia genome and 18 from B. socotrana (Fig. 2j).
Chromosome discrimination in Begonia hybrids. The digoxigenin-11-dUTP labeled tuberous Begonia genomic DNA was detected with anti-Dig FITC (pale/green fluorescence) and B. socotrana chromosomes were counterstained with DAPI (dark/blue fluorescence). a HO-7256-3 (2n = 28); b HO-7256-4 (2n = 28); c H08-124-01 (2n = 28); d ‘Netja Dark’ (2n = 42); e H07-102-13 (2n = 42); f H07-III-02 (2n = 41); g H08-143-03 (2n = 42); h 04-187-23 (2n = 63); i H08-132-01 (2n = 56); j H08-143-01 (2n = 45); k 04-122-04 (2n = 44); l 04-117-04 (2n = 56). Arrows indicate chromosomes carrying satellites. Bars = 5 μm (Color figure online)
For some genotypes with less condensed metaphases tuberous Begonia chromosomes with satellites were observed (Fig. 2, indicated by arrows). Recombinant chromosomes were not detected via GISH in any ‘Elatior’-begonia hybrids analyzed.
Discussion
Karyomorphology of ‘Elatior’-begonia hybrids
Begonia is characterized by small and morphologically little differentiated chromosomes. In this study the position of the centromeres was not discernible for B. socotrana chromosomes whereas in chromosomes derived from tuberous Begonias centromeres were distinct only for a few chromosomes at less condensed stage in prometaphase. Similar conclusions were drawn by Arends (1970) for ‘Elatior’-begonias and Okuno and Nagai (1954) and Legro and Haegeman (1971) for hybrids tuberous Begonias (B. × tuberhybrida Voss). The chromosomes of B. socotrana differ in size from those of tuberous Begonias, so it seems possible to identify them in ‘Elatior’-begonias hybrids. However, in our study, in a few metaphase plates chromosomes carrying a satellite were observed (Fig. 2. arrows) and the minute chromosomes of B. socotrana and satellites were difficult to recognize from each other in crowded metaphases of ‘Elatior’-begonias. Chromosomes carrying satellites were also observed for other species e.g., in B. evansiana (Okuno and Nagai 1953) and B. semperflorens (Zeilinga 1962).
Genome differentiation by GISH
GISH distinguished chromosomes derived from tuberous Begonia and B. socotrana in ‘Elatior’-begonia. The uniform labeling of tuberous Begonia genome was observed when the probe concentration was ~3.75 ng/μl and the ratio of probe to B. socotrana blocking DNA was 1:30. B. socotrana DNA used as a probe failed to label B. socotrana chromosomes both in presence and lack of the blocking DNA, which may be caused by the very small genome size-0.63 pg/1C (Marie and Brown 1993) and small amount of repetitive DNA. The problem with hybridization of genomic probes has been also observed in other plant species with small genomes. For many species with small genomes having a relatively low proportion of the middle and high repetitive DNA families, GISH signals were limited to the pericentromeric regions for example in Brassica (Hasterok et al. 2005), Oryza (Li et al. 2001), Rubus (Lim et al. 1998) and Brachypodium distachyon (Hasterok et al. 2004). However, there are also a few species with small genomes, e.g. in the genera Musa (D’Hont et al. 2000), Solanum and Lycopersicon (Gavrilenko et al. 2001) where genomic probes hybridize to the entire length of the chromosomes. This has been interpreted to be the result of a more even distribution of repetitive DNA families along the chromosomes. According to Raina and Rani (2001) the critical genome size below which it is difficult to paint along entire chromosomes might be around 0.6 pg/1C. Nevertheless, in Arabidopsis thaliana representing the smallest genomes among angiosperms (1C = 0.16 pg; according to Bennett et al. 2003) a uniform labeling of entire chromosomes was achieved by increasing DNA probe concentration up to 7.5–15 μg per slide or 5 μg of probe and increasing time of hybridization to 60 h (Ali et al. 2004). Due to satisfying differentiation of genomes in ‘Elatior’-begonia hybrids using single-target GISH with tuberous Begonia probe we did not try to modify the method by increasing concentration of B. socotrana DNA.
Chromosome constitutions of ‘Elatior’-begonia hybrids
Most of the progeny of ‘Elatior’-begonias are said to be triploids having 40–42 chromosomes, a few are diploids and occasionally tetraploids with 56 chromosomes (Mikkelsen 1976; Hvoslef-Eide and Munster 2006). In our study, seven different chromosome numbers have been found in eleven genotypes of ‘Elatior’-begonia hybrids (Table 2). The observation of 14 chromosomes of tuberous Begonia and 14 of B. socotrana in three accessions of ‘Elatior’-begonia hybrids (Fig. 2a–c) indicates that they are the result of hybridizing at diploid level of B. socotrana (2n = 28) with tuberhybirds (2n = 28). According to Arends (1971), crosses between diploid tuberous Begonia hybrids and B. socotrana do not succeed. However, Doorenbos (1973) obtained diploid ‘Elatior’-begonias hybrids from hybridization between diploid cultivar of the ‘Pendula’-type with B. socotrana.
Four ‘Elatior’-begonias hybrids investigated in our study (‘Netja Dark’, H07-102-13, HO7-III-02, H08-143-03) were triploids (27-28T + 14S) (Fig. 2d–g) whereas genotype H08-143-01 was near triploid (3x + 4) comprising of 27 tuberous Begonia and 18 B. socotrana chromosomes (Fig. 2j). Similar genome composition has been observed by Arends (1970) in other twenty-one ‘Elatior’-begonias hybrids. Our conclusion on the origin of these hybrids are consistent with those presented by Arends (1970) that they resulted from crosses of tuberhybirds with 2n = 54 or 56 chromosomes and B. socotrana with 2n = 28 chromosomes.
The hybrid HO8-132-01 (2n = 28T + 28S) (Fig. 2i) may be the result of backcrossing of ‘Elatior’-begonia (2n = 28T + 14S) with B. socotrana (2n = 28S) where the former genotype produced diploid egg cell. Similar genome constitution of ‘Elatior’-begonia can be expected from crosses between tetraploid tuberous Begonia hybrids (2n = 4x = 56) with the B. socotrana where the species provided unreduced gametes. Dewitte et al. (2009) proved that occurrence of viable 2n pollen is not a rare phenomenon in Begonia. In their study, 14% of investigated plants produced unreduced gametes. They have obtained successful crosses with genotypes producing 2n pollen, and showed via flow cytometry analysis the increase of the DNA content in the progenies. Similarly, in Begonia, allotetraploid semperflorens begonias have originated with the use of unreduced gametes (Horn 2004). However, there is no data on gamete formation in B. socotrana.
In genotype 04-122-04 (2n = 44) (Fig. 2k) all chromosomes showed uniform labelling with tuberous Begonia DNA probe. This plant has morphological characters of tuberous Begonia (date not shown) and may have developed apomictically from unfertilised maternal cells. After crosses between B. socotrana and B. × semperflorens-cultorum, Preil and Lorenz (1983) obtained progenies having the genotype of the male parent which must have originated from androgenesis or by elimination of the maternal chromosomes. Another possible explanation of genome composition in 04-122-04 may be elimination of B. socotrana chromosomes. The elimination of B. socotrana chromosomes has been suggested before by Arends (1970) for two ‘Elatior’-begonias ‘Riegers Leuchtfeuer’ (28T + 9S) and ‘Rose Queen’ (28T + 12S). Chromosome elimination has been reported in cross in other species, e.g. Nicotiana tabacum × N. plumbaginifolia (Ar-Rushdi 1957). The selective elimination of the chromosomes of one of the parental genotypes may be associated with disturbances in nuclear division (such as non-congressed chromosome at metaphase, lagging chromosomes and bridges at anaphase, chromatin fragments, degraded type of chromatin, multipolar spindles and micronuclei) in hybrid embryos and endosperm (Lange 1971; Bennett et al. 1976). According to Noda and Kasha (1981) the failure of congregation of some chromosomes during prometaphase appears to be main mitotic disturbance leading to chromosome elimination in hybrids between Hordeum vulgare and H. bulbosum. Gupta (1969) suggested that significant difference between the S and G2 periods of the parental species might lead to elimination of a chromosome segments or the whole genome of the species having the longer S or G2 phases.
In genotype 04-187-23 (Fig. 2h) 56 chromosomes were derived from tuberous Begonia and 7 chromosomes from B. socotrana. The same genome constitution was observed by Arends (1970) in a second generation ‘Elatior’-begonia ‘Eveleens Orange’ which originated as the result of crossing ‘Elatior’-begonia ‘Flambeau’ (2n = 52T + 14S) with the tuberous hybrid ‘Flamboyant’ (2n = 42T).
Basic chromosome number
The genome constitutions of ‘Elatior’-begonias 04-187-23 and ‘Eveleens Orange’ having 7 chromosomes of B. socotrana are difficult to explain considering the basic chromosome number in Begonia x = 13 and 14 which have been postulated by Legro and Haegeman (1971). It is also difficult to explain that according to Legro and Haegeman (1971) a triploid tuberous begonia ‘Tasso’ is propagated by seeds. These data may support suggestion of Matsuura and Okuno (1936, 1943) that the basic chromosome number in the genus Begonia is 6, 7 and 13, where the last may be of secondary origin derived from synthesis the former two. Similar conclusions were drawn by Okuno and Nagai (1953, 1954) based on analysis of meiotic chromosome configuration in B. evansiana and B. tuberohybrida. In our study, taking into consideration 6 and 7 as the basic chromosomes number in genus Begonia, B. socotrana (2n = 28) would be an autopolyploid which would explain genome composition in 04-187-23 and ‘Eveleens Orange’ (2n = 56T + 7S) with 7 chromosomes of B. socotrana. In Table 2 the ploidy level of analyzed genotypes were evaluated based on basic chromosome number x = 13 and 14 according to Legro and Haegeman (1971) and x = 6 and 7 according to Matsuura and Okuno (1936). This knowledge on the ploidy level and genome composition of analyzed ‘Elatior’-begonias is essential for further progress in breeding Begonias.
References
Ali HBM, Lysak MA, Shubert I (2004) Genomic in situ hybridisation in plants with small genomes is feasible and elucidated the chromosomal parentage in interspecific Arabidopsis hybrids. Genome 47:954–960
Aljanabi SM, Martinez I (1997) Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucleic Acids Res 25:4692–4693
Arends JC (1970) Somatic chromosome numbers in ‘Elatior’-begonias. Meded Landbouwh Wageningen 70–20:1–18
Ar-Rushdi AH (1957) The cytogenetics of variegation in a species hybrid in Nicotiana. Genetics 42:312–325
Barba-Gonzalez R, Van Silfhout AA, Ramanna MS, Visser RGF, Van Tuyl JM (2006) Progenies of allotriploids of Oriental × Asiatic lilies (Lilium) examined by GISH analysis. Euphytica 151:243–250
Bennett MD, Finch RA, Barclay IR (1976) The time, rate and mechanism of chromosome elimination in Hordeum hybrids. Chromosoma (Berl.) 54:175–200
Bennett MD, Leitch IJ, Hanson L (2003) DNA amounts in two samples of angiosperm weeds. Ann Bot 82(Supplement A):121–134
D’Hont A, Paget-Goy A, Escoute J, Carreel F (2000) The interspecific genome structure of cultivated banana. Musa spp., revealed by genomic DNA in situ hybridization. Theor Appl Genet 100:177–183
Dewitte A, Eckhaut T, van Huylenbroeck J, van Bockstaele E (2009) Occurrence of viable unreduced pollen in a Begonia collection. Euphytica 161:81–94
Doorenbos J (1973) Breeding ‘Elatior’-begnia (B. × hiemalis Fotsch). Acta Hort 31:127–131
Doorenbos J, Legro RAH (1968) Breeding ‘Gloire de Lorraine’ Begonias. Meded Landbouwh Wageningen 68:19
Doorenbos J, Sosef MSM, de Wilde JJFE (1998) The section of Begonia. Wageningen Agricultural University, Wageningen, The Netherlands
Fulton MT, Chunzoongse J, Tanksley SD (1995) Microprep protocol for extraction of DNA from tomato and other herbaceous plants. PMB Rep 13:207–209
Gavrilenko T, Thieme R, Rokka VM (2001) Cytogenetic analysis of Lycopersicon esculentum (+) Solanum etuberosum somatic hybrids and their androgenetic regenerants. Theor Appl Genet 103:231–239
Gleed CJ (1961) Hybrid winter-flowering Begonias. The story of raising some early varieties. J R Hort Soc 86:319–322
Gupta SB (1969) Duration of mitotic cycle and regulation of DNA replication in Nicotiana plumbaginifolia and a hybrid derivative of N. tabacum showing chromosome instability. Can J Genet Cytol 11:133–142
Haider Ali SN, Ramanna MS, Jacobsen E, Visser RGF (2002) Genome differentiation between Lycopersicon esculentum and L. pennellii as revealed by genomic in situ hybrization. Euphytica 127:227–234
Hasterok R, Jenkins G, Longdon T, Jones RN, Maluszynska J (2001) Ribosomal DNA is an effective marker of Brassica chromosomes. Theor Appl Genet 103:486–490
Hasterok R, Draper J, Jenkins G (2004) Laying the cytotaxonomic foundations of a new model grass. Brachypodium distachyon (L.) Beauv. Chromosome Res 12:397–403
Hasterok R, Wolny E, Kulak S, Zdziechiewicz A, Maluszynska J, Heneen WK (2005) Molecular sytogenetic analysis of Brassica rapa-Brassica oleracea var. alboglabra monosimic additon liens. Theor Appl Genet 111:196–205
Horn W (2004) The patterns of evolution and ornamental plant breeding. Acta Hort 651:19–31
Hvoslef-Eide AK, Munster C (2006) Begonia. History and breeding. In: Anderson NO (ed) Flower Breed Genet, Chapter 9. Springer, Heidelberg, pp 241–275
Kamstra SA, Kuipers AGJ, De Jeu MJ, Rammana MS, Jacobsen E (1999) The extent and position of homoelogous recombination in a distant hybrid of Alstroemeria: a molecular cytogenetic assessment of first generation backcross progenies. Chromosoma 108:52–63
Kopperud C, Einset JW (1995) DNA isolation from begonia leaves. PMB Rep 13:129–130
Kroon GH (1993) Breeding research in Begonia. Acta Hort 337:53–58
Lange W (1971) Crosses between Hordeum vulgare L. and H. bulbosum L. II. Elimination of chromosomes in hybrid tissue. Euphytica 20:181–194
Legro RAH, Doorenbos J (1969) Chromosome number in Begonia. Neth J Agric Sci 17:189–202
Legro RAH, Haegeman JFV (1971) Chromosome number of hybrid tuberous Begonias. Euphytica 20:1–13
Li CB, Zhang DM, Ge S, Lu BR, Hong DY (2001) Differentiation and inter-genomic relationships among C, E and D genomes in the Oryza officinalis complex (Poaceae) as revealed by multicolour genomic in situ hybridization. Theor Appl Genet 103:197–203
Lim KY, Leitch IJ, Leitch AR (1998) Genomic characterization and the detection of raspberry chromatin in polyploid Rubus. Theor Appl Genet 97:1027–1033
Marasek A, Mizuochi H, Okazaki K (2006) The origin of Darwin hybrid tulips analyzed by flow cytometry, karyotype analyses and genomic in situ hybridization. Euphytica 151:279–290
Marie D, Brown SC (1993) A cytometric exercise in plant DNA histograms, with 2C values for 70 species. Biol Cell 78:41–51
Matsuura H, Okuno S (1936) Cytological study in Begonia. Jpn J Genet 12:42–43
Matsuura H, Okuno S (1943) Cytological study on Begonia (Preliminary survey). Cytologia 13:1–18
Mikkelsen JC (1976) Enlarging the scope of flower crop improvement mutation breeding. Acta Hort 63:197–202
Murray MG, Thompson WF (1980) Rapid isolation of high-molecular-weight plant DNA. Nucleic Acid Res 8:4321–4325
Nakata M, Guan K, Li J, Lu Y, Li H (2007) Cytotaconomy of Begonia rubropunctata and B. purpureofolia (Begoniaceae). Bot J Lin Soc 155:513–517
Noda K, Kasha KJ (1981) Chromosome elimination on different meristematic regions of hybrids between Hordeum vulgare L. and H. bulbosum L. Jpn J Genet 56:193–204
Okuno S, Nagai S (1953) Cytological studies on Begonia Evansiana Andr. with special reference to its meiotic chromosomes. Jpn J Genet 28:132–136
Okuno S, Nagai S (1954) Karyotypic polymorphism in Begonia tuberohybrida Voss. Jpn J Genet 29:185–196
Preil W, Lorenz A (1983) Über das Auftreten matromorpher, patromorpher und intermediärer Nachkommen nach Kreuzungen zwischen Begonia socotrana (Hook.) und Begonia × semperflorens-cultorum. Z. Pflanzenzücht. 91: 253–260
Raina SN, Rani V (2001) GISH technology in plant genome research. Methods Cell Sci 23:83–104
Wagner WW (1999) The French begonia society. The Begonian 66:172–175
Ye HG, Wang FG, Ye YS, Peng CI (2004) Begonia coptidifolia (Begoniaceae), a new species form China. Bot Bull Acad Sin 45:259–266
Zeilinga AE (1962) Cytological investigation of hybrid varieties of Begonia semperflorens. Euphytica 11:126–136
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Marasek-Ciolakowska, A., Ramanna, M.S., ter Laak, W.A. et al. Genome composition of ‘Elatior’-begonias hybrids analyzed by genomic in situ hybridisation. Euphytica 171, 273–282 (2010). https://doi.org/10.1007/s10681-009-0057-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-009-0057-7