Abstract
The fine-scale genetic structure and how it varies between generations depends on the spatial scale of gene dispersal and other fundamental aspects of species’ biology, such as the mating system. Such knowledge is crucial for the design of genetic conservation strategies. This is particularly relevant for species that are increasingly fragmented such as Boswellia papyrifera. This species occurs in dry tropical forests from Ethiopia, Eritrea and Sudan and is an important source of frankincense, a highly valued aromatic resin obtained from the bark of the tree. This study assessed the genetic diversity and fine-scale spatial genetic structure (FSGS) of two cohorts (adults and seedlings) from two populations (Guba-Arenja and Kurmuk) in Western Ethiopia and inferred intra-population gene dispersal in the species, using microsatellite markers. The expected heterozygosity (H E) was 0.664–0.724. The spatial analyses based on kinship coefficient (F ij) revealed a significant positive genetic correlation up to a distance of 130 m. Spatial genetic structure was relatively weak (Sp = 0.002–0.014) indicating that gene dispersal is extensive within the populations. Based on the FSGS patterns found, we estimate indirectly gene dispersal distances of 103 and 124 m for the two populations studied. The high heterozygosity, the low fixation index and the low Sp values found in this study are consistent with outcrossing as the (predominant) mating system in B. papyrifera. We suggest that seed collection for ex situ conservation and reforestation programmes of B. papyrifera should use trees separated by distances of at least 100 m but preferably 150 m to limit genetic relatedness among seeds from different trees.
Similar content being viewed by others
Introduction
The spatial genetic structure (SGS) of populations is determined by various processes including gene flow and local selection at different life history stages. In plant species, gene dispersal is mediated by pollen and seed and along with other factors, it influences how genetic diversity is structured within and between populations. Seed movement allows migration and range expansion, while the spatial scale of gene flow via both seed and pollen has important implications for the maintenance of genetic diversity (Peakall et al. 2003; Moran and Clark 2011). Limited dispersal will enhance population differentiation, and in the long term, it may hinder the ability of a species to colonize new sites or to shift the range in response to environmental changes (Moran and Clark 2012).
At a fine spatial scale, the genetic similarity is, in general, higher among neighbouring than among more distant individuals. A local genetic structure can develop due to limited gene flow, generally when seed-mediated gene flow is low even in a large continuous population (Vekemans and Hardy 2004; Segelbacher et al. 2010; Moran and Clark 2012). Moreover, life form (e.g. annual or perennial), density of the population, and the breeding system of species are important determinants of intra-population spatial genetic structure (Duminil et al. 2009). Due to limited gene flow and the effects of these factors, genetically related individuals aggregate, producing fine-scale spatial genetic structure (FSGS), which is a decrease of pairwise kinship with increasing pairwise distance (Vekemans and Hardy 2004). The structure is reinforced in subsequent generations due to bi-parental inbreeding and genetic drift if pollen movement and seed dispersal are limited (King and Zalucki 2012; Moran and Clark 2012). On the other hand, extensive dispersal and immigration would weaken FSGS over generations. Therefore, investigating the pattern of FSGS and how it varies between generations provides insight into the intensity and spatial scale of gene dispersal and other fundamental aspects of species’ biology, such as the mating system (Segelbacher et al. 2010) or clonality (Dering et al. 2016; Jankowska-Wroblewska et al. 2016). Knowledge of the scale of gene dispersal within populations of species is a crucial information for the design of a conservation strategy that will be effective for the maintenance of genetic diversity of the species.
Boswellia papyrifera grows in dry tropical forests, and it is threatened across the whole distribution area. In many areas in Ethiopia, the natural stands of B. papyrifera now consist of fragmented patches of trees because of land-use changes, grazing, fire and adult mortality. Populations in Northern (Abergele) and Northwestern (Metema) Ethiopia are suffering from lack of saplings and small trees (Abiyu et al. 2010; Eshete et al. 2011; Groenendijk et al. 2012; Tolera et al. 2013). In these Boswellia populations, non-permanent seedlings (seedlings of which the shoots die back during the dry season and reappear in the rainy season) are abundant, but they do not develop to the sapling stage mainly because of the impacts of frequent forest fire and intensive grazing in the area (Groenendijk et al. 2012; Tolera et al. 2013). A similar situation was reported for Eritrea (Ogbazghi et al. 2006; Rijkers et al. 2006) and Sudan (Adam and El Tayeb 2008; Abtew et al. 2012). Boswellia sacra in Oman (Farah 2008) and both Boswellia serrata (Sunnichan et al. 2005) and Boswellia ovalifoliolata (Raju et al. 2012) populations in India also show regeneration problems.
During our fieldwork in Western Ethiopia, we discovered B. papyrifera populations that did have a young cohort of seedlings and saplings next to the mature, flowering trees (Addisalem et al. 2016). These unique, naturally regenerating populations enabled us to study the FSGS of the species, for seedling and adult cohorts. Our study aimed at assessing (i) the differences in genetic diversity between the parent and progeny cohorts, (ii) the FSGS within the two cohorts in each of two populations and (iii) the gene dispersal distances from FSGS patterns. The results are discussed in view of the urgent need to define strategies of conservation for this emblematic species.
Materials and methods
Study species
B. papyrifera (Del.) Hochst. (Burseraceae) occurs in Terminalia-Combretum dry tropical forests. The species is an important source of frankincense, a highly valued aromatic resin obtained from the bark of the trees. Due to its use in the production of perfumes and pharmaceuticals, frankincense is internationally traded and is one of the export commodities of Ethiopia, Sudan and Eritrea (Coppen 2005; Lemenih 2005; Ogbazghi et al. 2006; Abtew et al. 2012). In Ethiopia, B. papyrifera tree stands occur mainly in the North, Northeast, Northwest and Western parts of the country (Eshete et al. 2011; Girma et al. 2013) and in Combretum-Terminalia and Acacia-Commiphora woodlands. B. papyrifera is a widespread, slow growing and long-lived (>100 years) species (Tolera et al., 2013). It is a deciduous tree, monoecious with sweet-scented flowers, frequently visited by honeybees for pollen and nectar (Fichtl and Admasu 1994), indicating honeybees as one of its pollinators. Two closely related species, B. serrata and B. ovalifoliolata are also insect pollinated (Sunnichan et al. 2005; Raju et al. 2012). The fruit of B. papyrifera, a drupe, is about 2 cm long and usually contains three tapered seeds (Vollesen 1989). The seed dispersal mode of B. papyrifera is not well known. Seeds of B. ovalifoliolata disseminate up to a distance of 400 m (Raju et al. 2012).
Study area and sampling
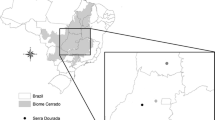
The study was conducted in the Western part of Ethiopia, which is the only place, as far as we know, in the species’ distribution range in Ethiopia, where B. papyrifera regeneration and stand recruitment amply occurs (Addisalem et al. 2016). After a preliminary survey of the distribution of the species within the region, two populations, Guba-Arenja located at 11.05° N, 35.16° E and Kurmuk (10.57° N, 34.30° E) (Fig. 1), were selected for this study. Both are continuous populations, and they were completely accessible for sampling. The two populations were treated as replications for the FSGS analysis of the species. All four life cycles/size classes described in Groenendijk et al. (2012) were present simultaneously in these two populations, although the relative abundance differed between the two populations.
In Guba-Arenja, the sampled area was a strip running along a slope from the top of a hill (highest altitude = 764 m a.s.l.) to downhill (lowest altitude = 720 m), where it was embedded with a seasonal river course. It contained a patchily distributed population of B. papyrifera with intermittent grasslands. Recently, a highway had been constructed across the slope. The population was characterized by patches of abundant seedlings and saplings and a few small and big-sized adult trees. Unlike Groenendijk et al. (2012) in the population in Metema, in the North of Ethiopia, we observed that several small B. papyrifera individuals as tall as only 1.5 m were already flowering in this population.
The Kurmuk population was located in a relatively flat area with altitude 662–680 m a.s.l. surrounded by a hilly area in the West and Northwest. The B. papyrifera population in this area was characterized by the presence of abundant seedlings, saplings, small trees, and a few scattered big adult trees. Individuals were uniformly distributed across the population.
The pattern of FSGS of two life stages, adult and seedling cohorts, was investigated within each population. For the purpose of this study, we considered all trees with DBH >10 cm as ‘adults’ presumably capable of producing pollen and seeds, and they were all considered as potential parents. Our ‘seedling’ cohort consisted of small plants up to a height of ~50 cm. These ‘seedlings’ are the youngest plants present in the population, but they may be up to several years old already (Birhane et al. unpublished data). Their height distinguishes them from larger saplings and juvenile trees and allowed us to get the largest contrast in plant age and developmental stage possible within the populations.
Sampling individuals and plant material
We sampled 106 adult trees and 105 seedlings from Guba-Arenja (22-ha plot) and 80 adult trees and 60 seedlings from Kurmuk (36-ha plot). The difference in the number of samples was due to the limitation of trees bearing young leaves during the sample collection, as we only sampled trees and seedlings with young leaves. Samples of young leaves were immediately stored in silica gel, which was regenerated regularly until the samples were dry. All samples were georeferenced using a Garmin Dakota 20 GPS (which is accurate to within 15 m, according to Garmin). As the adult density was 58 trees per hectare in Guba-Arenja and 87 trees per hectare in Kurmuk (Teshome 2013; Teshome et al., unpublished data), our sample of adult trees represented about 8 % (Guba-Arenja) and 3.5 % (Kurkum) of all adult trees. The sampled individuals were randomly distributed across the populations; therefore, we assume that the sample is spatially representative.
DNA extraction
Total DNA was isolated from dried young leaves following the cetyl trimethylammonium bromide (CTAB) protocol of Fulton et al. (1995). As large amounts of phenolic compounds were expected because of the resin content in the leaves, the protocol was modified by the addition of 2 % pvp-40 in the extraction buffer and 1 % mercaptoethanol in the microprep buffer following the methods described in Fulton et al. (1995). Purification was performed using DNeasy (Qiagen, Venlo, The Netherlands) procedures following Smulders et al. (2010). DNA yield and quality were visually assessed on 1 % agarose gels.
Genotyping
The samples were genotyped using six polymorphic microsatellite loci, namely Bp17, Bp20, Bp21, Bp22, Bp23 and Bp39 (Table 1), developed previously for this species (Addisalem et al. 2015). The loci were selected based on their level of polymorphism in terms of the total number of alleles (10–28 alleles per locus) in a previous population structure study (Addisalem et al. 2016) that included 12 populations across Ethiopia. Descriptions of the primers, the PCR mix, labelling of the primers and the thermal cycling profile of the PCRs are detailed in Addisalem et al. (2015). Forward primers were labelled with a IRD700 or IRD800 fluorescent dye. After amplification, the mixture was diluted with 10 μl water and 5–6 μl of fluorescently labelled PCR products resolved on a 4200 or 4300 LI-COR DNA analyzer. The gel was reloaded once or twice, and if the resolution was insufficient, the PCR products were (re)analysed on a new gel. Alleles were scored manually. The length of the alleles was estimated using SequaMark 10 bp DNA step ladder.
Genetic diversity analysis
Genetic diversity was estimated separately for the two populations and for the two cohorts (adults and seedlings). The total number of alleles (A), the effective number of alleles (A E), the observed (H O) and expected heterozygosity (H E) and the fixation index (F) were calculated for each population and each cohort using GenAlEx 6.5 (Peakall and Smouse 2006, 2012). Genetic differentiation between the populations from the two sites (seedling and adult cohorts combined) and between cohorts within each population (adults versus seedlings) was analysed based on F ST estimates.
Fine-scale spatial genetic structure
FSGS can be described by computing pairwise relatedness coefficients between individuals in the sample and analysing their relationship with the spatial distance separating the individuals (Vekemans and Hardy 2004). In this study, we characterized FSGS using the kinship relatedness coefficient (F ij) (Hardy and Vekemans 1999) and the autocorrelation coefficient (r) (Peakall et al. 2003). Analyses were based upon genetic and geographic distance matrices calculated using data from the six microsatellite markers and field GPS records of the plants sampled. Distance class sizes (20, 40, 80, 160, 300, 500 and 850 m) were specified such that the first distance interval would calculate relatedness coefficients based on all pairwise comparisons within a distance of 0–20 m, the second analysis for 20–40 m, and so on until the total spatial distance was covered. We used the same distance classes in both populations and both cohorts to be able to compare the results between the two sites and between the two cohorts within a population. However, as the number of pairs of individuals was too low at the first interval distance (20 m) in the Kurmuk population and at the last distance interval (850 m) in the Guba population, these distance intervals were not considered.
The pattern of FSGS based on the kinship or co-ancestry coefficient (F ij), i.e. the probability that a random gene from individual i is identical to a random gene from individual j (Vekemans and Hardy 2004), was analysed using SPAGeDi 1.4b (Hardy and Vekemans 2002). The slope of regression (b̂ F ) (kinship coefficient regressed over logarithmic geographic distance) and the historical gene dispersal parameter (σ) were estimated using the same programme. The significance of the regression slope b̂ F was tested against the null hypothesis (the overall absence of FSGS, b̂ F = 0) by comparing the observed values with those obtained after 1000 random permutations of individuals among positions. The extent of the FSGS was quantified using the Sp statistic (Sp), calculated as −b̂ F /(1 − F̂ 1) described in Vekemans and Hardy (2004), where b̂ F is the regression slope and F̂ 1 is the mean F ij between individuals belonging to the first distance class containing adequate pairs of individuals to precisely estimate the F ij. The neighbourhood size and the gene dispersal distance σ g were jointly estimated for each population relying on SGS patterns following the iterative procedure described in Hardy et al. (2006). The method posits that F ij is expected to decay linearly with the ln(distance) at a rate inversely proportional to the product D E·σg 2 for a distance range between σ g and ca. 20 σ g, where D E is the effective density of reproductive individuals and σ g 2 is the axial variance of gene dispersal distance between two generations. The joint estimate is sensible to the quality of D E (Hardy et al. 2006). Accordingly, different values of D E were tested, considering one half, one fourth and one tenth of the adult densities to account for the lifetime variation in reproductive success among adult trees (Hardy et al. 2006).
The pattern of FSGS is affected by the mating system, gene dispersal as well as ecological factors; hence, different situations can be expected to yield different autocorrelation patterns. We applied the nonparametric heterogeneity test described by Smouse et al. (2008), implemented in GenAlEx 6.5, to test the statistical significance of the spatial autocorrelation patterns (analogous to the pattern based on kinship coefficient) observed between the two populations and the two cohorts within each population. The method for calculating the autocorrelation coefficient (r) is described in Peakall and Smouse (2006, 2012). The statistical significance of r was determined using 999 permutations, randomizing genotypes among distance classes. Within a specific distance class, spatial genetic structure was considered significant when the observed r value fell outside of the 95 % CI and when the error bar did not intersect with r = 0 (Peakall and Smouse 2006, 2012). Single-class (t 2) and multi-class test statistic (ω) were computed with 999 permutations.
Results
Genetic diversity
Across the six microsatellite loci and the 330 genotypes (adults and seedlings in both populations), 98 alleles were detected in total, with an average of 16.3 alleles per locus. The number of alleles per locus (A) ranged from 5 (at locus Bp39) to 30 (at locus Bp22). The common alleles in these populations were also found in our previous study that included 12 populations from across Ethiopia (Addisalem et al. 2016), but more alleles were detected for some of the loci (e.g. 30 alleles at Bp22) and fewer for other loci (e.g. 5 alleles at Bp39). Mean H O and H E across all cohorts and across the two populations were 0.689 and 0.693, respectively (Table 1). Genetic differentiation between the two study sites, Guba-Arenja and Kurmuk (adult and seedling cohorts in each population combined), was low (F ST = 0.038).
Genetic diversity between generations/cohorts
The number of alleles was the same in both cohorts within each of the populations (Table 2). In Guba-Arenja, the total number (A) was 14 and the effective number (A E) was 7 in both adult (GA) and seedling (GS) cohorts. In Kurmuk, slightly fewer alleles (A = 10 and A E = 4) were detected in both cohorts. Heterozygosity was 0.707 and 0.724, respectively, in adult (GA) and seedling (GS) cohorts in Guba-Arenja and 0.664 and 0.677 in adult (KA) and seedling (KS) cohorts in Kurmuk (Table 2). The F ST between the adult and seedling cohorts were 0.010 in Guba-Arenja and 0.004 in Kurmuk population.
Fine-scale spatial genetic structure
The kinship coefficient (F ij) and the autocorrelation coefficient (r) showed a similar FSGS pattern. Here, we will first present the SPAGeDi results on kinship and the extent of SGS (Sp) analysis in detail, and then report the results from GenAlEx on the cross-population heterogeneity test.
An overall positive signal of local spatial genetic structure was detected in the two B. papyrifera populations except in the Kurmuk seedling cohort. Significant genetic relatedness (F ij) was detected up to ca. 130, 100 and 60 m, respectively, in Guba-Arenja adult (GA), Guba-Arenja seedling (GS) and Kurmuk adult (KA) cohorts (Fig. 2). In Kurmuk, the seedling cohort (KS) F ij was not significant over the whole distance range (Fig. 2). We also analysed adult and seedling cohorts combined in each population (GC and KC) for comparison at landscape level. The results showed significant positive SGS up to a distance of ca. 100 m in GC (Fig. 2) and no significant relatedness in KC (Fig. 2).
The kinship coefficient (F ij) plotted against geographical distances (in meters). Dashed lines represent 95 % confidence intervals for F ij under the null hypothesis that genotypes are randomly distributed. Vertical bars are standard errors of means. (Upper left) Guba-Arenja adult cohort; (central left) Guba-Arenja seedling cohort; (lower left) Guba-Arenja seedling and adult cohorts combined; (upper right) Kurmuk adult cohorts; (central right) Kurmuk seedling cohorts; (lower right) Kurmuk seedling and adult cohorts combined
The kinship coefficient over the shortest distance (F 1) was 0.044 in GA and GS. In Kurmuk, the estimates were 0.044 for KA and 0.004 for KS (Table 3). The regression slope (b̂ F ) of the mean kinship coefficients on the logarithm of spatial distance was negative and significant in GA, GS and KA and not significant in KS. The statistic for the extent of the spatial genetic structure (Sp) was 0.014 ± 0.005 (SE) in GA and 0.011 ± 0.0034 in KA. Estimated historical gene dispersal parameters (σ) based on adults and assuming effective densities (D E) equal to D/2, D/4 and D/10 were, respectively, 46 ± 11.5, 64 ± 14 and 103 m for Guba-Arenja and 65 ± 25, 81 ± 25, and 124 m for Kurmuk populations (Table 3).
Heterogeneity analyses of the FSGS
The spatial autocorrelation analysis revealed a similar pattern of autocorrelation to the kinship analysis over similar distances in each of the cohorts and populations. The test for cross-population heterogeneity based on the single-class test (t 2) and multi-class criterion (ω) showed that the pattern of spatial genetic structure did not significantly differ between the two populations and between the two cohorts within each of the populations (Supplementary Table 1). The test between populations considering only adult cohorts also did not show a significant difference.
Discussion
Genetic diversity between generations/cohorts
We used six microsatellite markers to determine the genetic diversity and FSGS of two populations of B. papyrifera in Western Ethiopia. Genetic diversity was high both in terms of the number of alleles and heterozygosity. The level of genetic diversity was similar in adult and seedling cohorts in both Guba-Arenja and Kurmuk populations. The high mean number of alleles detected per cohort (10–14) suggests a large effective population size. The level of heterozygosity detected in these two populations (H O = 0.689, H E = 0.693) was similar to results observed in other tropical tree species when measured with microsatellite markers (Acacia senegal (Omondi et al., 2010), Swietenia macrophylla (Lemes et al. 2003) and Parashorea stellata (Tiep 2015)). Acacia senegal is a tree species occurring in the same Terminalia-Combretum woodlands as B. papyrifera. We found low F ST between the two populations (F ST = 0.031). The two cohorts (adult versus seedling cohorts) in the two populations were not significantly differentiated.
The overall and effective number of alleles was slightly higher in Guba-Arenja than in Kurmuk. The number of alleles was consistently the same for adult and seedling cohorts while the heterozygosity was slightly higher in seedling cohorts of both populations indicating that the diversity in adult cohorts is adequately represented in their progeny (Table 2).
Fine-scale spatial genetic structure
The high degree of heterozygosity, the low fixation index and the low Sp values may indicate extensive intra-population gene dispersal. Sp in predominantly outcrossing or self-incompatible species is lower (average 0.0133) than in predominantly selfing species (average 0.143) (Hardy et al. 2006). The Sp values obtained here for B. papyrifera (0.011–0.014) are similar to the average of outcrossing species (0.013). The mating system of B. papyrifera has not been studied. However, two taxonomically close species are outcrossing (B. serrata —Sunnichan et al. (2005) and B. ovalifoliolata—Raju et al. (2012)). These species also show a pre-zygotic self-incompatibility mechanism (Sunnichan et al. 2005; Raju et al. 2012). The heterozygosity values we have obtained strongly suggest that outcrossing is the (predominant) mating system in B. papyrifera as well, and it may have a similar self-incompatibility system. In outcrossing species, pollen dispersal contributes to gene dispersal (Vekemans and Hardy 2004). Therefore, within-population gene dispersal detected in B. papyrifera may also be a result of the contribution of extensive pollen dispersal.
SGS may vary widely between populations (Jump et al. 2012), but comparing populations can be problematic because of differences in the sampling design. To avoid this problem, we have used the same strategy during field sampling, used the same microsatellite markers for the genetic analysis, and analysed SGS using the same distance classes. In this way, an overall positive signal of local spatial genetic structure was detected for both B. papyrifera populations except in the Kurmuk seedling cohort. From the kinship analyses, a significant genetic structure was also detected for both B. papyrifera populations, although in Kurmuk, no significant spatial genetic structure was detected for the seedlings over the whole distance range. Limited gene flow reinforces FSGS across generations (Smouse et al. 2008; King and Zalucki 2012; Moran and Clark 2012). In B. papyrifera, FSGS did not significantly change from adult to seedling populations. The absence of significant SGS in the seedling cohorts of Kurmuk may suggest that seed dispersal was efficient these last years in this population, whereas it was not across many seedling cohorts before (integrated into the current adult cohort). This may be due to an influx of seeds from the forest on a nearby slope, which appears to be more degraded (decreased tree density) and more open. Rainwater may cause surface soil runoff from the steep exposed area, taking the seeds with it. Indeed, very few seedlings were found at the top of the slope, while many seedlings were present in the flat sampled area which had deeper, moist and fertile soil, a conductive condition for germination of seeds and establishment of seedlings and saplings. Unfortunately, we do not know the history of deforestation in this area.
The intensity of FSGS in B. papyrifera (Sp = 0.011–0.013) was similar to that of three other tropical tree species, the wind-dispersed Dalbergia nigra (Sp = 0.017 (De Oliveira Buzatti et al. 2012)) and the bird-dispersed Protium spruceanum (mean Sp = 0.008 (Vieira et al. 2010)) and Pouteria reticulate (Sp = 0.006–0.010 (Schroeder et al. 2014)). All are insect-pollinated. In plant species, FSGS is related to life form, population density and mating system (Vekemans and Hardy 2004). In our study, the Guba-Arenja adult population was less dense (58 individuals/ha) than that in Kurmuk (87 individuals per hectare) but the Sp values were similar (0.014 and 0.011). Commonly (e.g. Chamaecrista fasciculata (Fenster 1991), Heliconia acuminata (Cortes et al. 2013)), higher gene dispersal distances tend to be found in low-density populations (Vekemans and Hardy 2004), although in mixed stands high tree density may limit dispersal by serving as a physical barrier to the movements of propagules (Curtu et al. 2015). Our estimated gene dispersal distance was 103 m in Guba-Arenja and 124 m in the denser population in Kurmuk. However, as we only used a small number of microsatellite markers, and only compared two populations, we cannot conclude whether density has an effect on gene dispersal in B. papyrifera in Western Ethiopia, and in which direction. For this, further studies of additional populations are needed, using a larger number of microsatellite loci.
The heterogeneity test for autocorrelation showed that SGS was not significantly different between populations or between cohorts. A long life span ensures the representation of many cohorts within a population. This enables such species to maintain diverse individuals across cohorts, ensuring a high genetic diversity within populations (King and Zalucki 2012; Moran and Clark 2012). As a long-lived, widespread species with extensive gene dispersal, B. papyrifera may maintain its genetic diversity across generations in continuous natural forest landscapes, but it may be at risk of losing the genetic diversity in the future in highly fragmented landscapes.
Conservation implications
This study provides the first assessment of FSGS and gene dispersal patterns in B. papyrifera. Such information can be valuable for management objectives that seek conservation of the genetic resources of the species. The high levels of heterozygosity and the low Sp values in the adult and seedling cohorts indicate that FSGS in B. papyrifera is relatively weak and gene dispersal is extensive within the populations. The absence of FSGS in one of the seedling cohort populations might be the consequence of seed deposit from sources originating outside of the sampled area. The current FSGS analysis provides information that may be used during the sampling of individuals and seed collection within conservation units for ex situ conservation and reforestation programmes. Our findings suggest that, within the wider conservation units/provenances, the collection of seeds for ex situ conservation and reforestation programmes of B. papyrifera should use trees separated by minimum distances of 100 but preferably 150 m to reduce genetic relatedness among seeds from different trees.
The B. papyrifera populations in Ethiopia are increasingly fragmented, and scattered remnant trees and populations dominate the growing region of the species. The species has maintained a high level of genetic diversity in adult trees in all populations studied (Addisalem et al. 2016). However, most of these populations do not have younger plants, preventing future regeneration. Although the findings of this study are important for managing and conserving the extant trees and populations in the fragmented landscapes, this study only provides baseline information on the spatial structuring and dispersal of genes in the species. Facilitating seed production and dispersal and survival of emerging seedlings are crucial for maintaining the populations viable and ensuring their sustainability. The fact that the Western populations of the present study do show high genetic diversity in combination with seedling recruitment is encouraging. If regeneration is made possible in other areas in Ethiopia, the genetic diversity may be conserved in all the current populations. Conservation measures thus should focus on the protection of current populations and on effective and long-lasting regeneration of populations.
References
Abiyu A, Bongers F, Eshete A, Gebrehiwot K, Kindu M, Limeneh M, Moges Y, Ogbazghi W, Sterck FJ (2010) Incense woodlands in Ethiopia and Eritrea: regeneration problems and restoration possibilities. In: Bongers F, Tenningkeit T (eds) Degraded forest in eastern Africa: management and restoration. Earthscan, London, UK
Abtew AA, Pretsch J, Mohamoud TE, Adam YO (2012) Population status of Boswellia papyrifera (Del.) Hochst in the dry woodlands of Nuba Mountains, South Kordofan State, Sudan. Agric For 54(8):41–50
Adam AA, El Tayeb AM (2008) A comparative study of natural regeneration on B. papyrifera and other tree species in Jebel Marra Darfur, Sudan. Res J Agric Biol Sci 4(1):94–102
Addisalem AB, Esselink GD, Bongers F, Smulders MJM (2015) Genomic sequencing and microsatellite marker development for Boswellia papyrifera, an economically important but threatened tree native to dry tropical forests. Aob Plants 7:plu086. doi:10.1093/aobpla/plu086
Addisalem AB, Bongers F, Kassahun T, Smulders MJM (2016) Genetic diversity and differentiation of the frankincense tree (Boswellia papyrifera (Del.) Hochst) across Ethiopia and implications for its conservation. For Ecol Managem 360:253–260. doi:10.1016/j.foreco.2015.10.038
Coppen JJW (2005). Overview of international trades and markets. In: Chikamai B and Casadei E (eds) Production and marketing of gum resins: frankincense, myrrh and opoponax. Network for Natural Gums and Resins in Africa (NGARA). Publication Series No. 5. NGARA, KEFRI, Nairobi, pp 5–34
Cortes MC, Uriarte M, Lemes MR, Gribel R, Kress WJ, Smouse PE, Bruna EM (2013) Low plant density enhances gene dispersal in the Amazonian understory herb Heliconia acuminata. Mol Ecol 22:5716–5729
Curtu AL, Craciunesc I, Enescu CM, Vidalis A, Sofletea N (2015) Fine-scale spatial genetic structure in a multi-oak-species (Quercus spp.) forest. iForest 8:324–332. doi:10.3832/ifor1150-007
De Oliveira Buzatti RS, Ribeiro RA, de Lemos Filho JP, Lovato MB (2012) Fine-scale spatial genetic structure of Dalbergia nigra (Fabaceae), a threatened and endemic tree of the Brazilian Atlantic Forest. Gen Mol Biol 35:838–846
Dering M, Rączka G, Szmyt J (2016) Sex-specific pattern of spatial genetic structure in dioecious and clonal tree species, Populus alba L. Tree Gen Genom 12:70. doi:10.1007/s11295-016-1028-5
Duminil J, Hardy OJ, Petit RJ (2009) Plant traits correlated with generation time directly affect inbreeding depression and mating system and indirectly genetic structure. BMC Evol Biol 9:177
Eshete A, Sterck FJ, Bongers F (2011) Diversity and production of Ethiopian dry woodlands explained by climate- and soil-stress gradients. For Ecol Managem 261:1499–1509
Farah MH (2008) Non-timber forest products (NTFP) extraction in arid environments: Landuse change, frankincense production and the sustainability of Boswellia sacra in Dhofaar (Oman). PhD Dissertation, University of Arizona. UMI number: 3315953
Fenster CB (1991) Gene flow in Chamaecrista fasciculata (Leguminosae). I Gene dispersal. Evolution 45:398–409
Fichtl R. Admasu A (1994) Honeybee flora of Ethiopia. Weikershem, Germany: Margraf Verlag pp 510
Fulton TM, Chunwangse J, Tanksley SD (1995) Microprep protocol for extraction of DNA from tomato and herbaceous plants. Plant Mol Biol Rep 13:207–209
Girma A, Skidmorea AK, de Biea CAJM, Bongers F, Schlerf M (2013) Photosynthetic bark: use of chlorophyll absorption continuum index to estimate Boswellia papyrifera bark chlorophyll content. Int J Appl Earth Obs 23:71–80
Groenendijk P, Eshete A, Sterck FJ, Zuidema PA, Bongers F (2012) Limitations to sustainable frankincense production: blocked regeneration, high adult mortality and declining populations. J Appl Ecol 49:164–173
Hardy OJ, Vekemans X (1999) Isolation by distance in a continuous population: reconciliation between spatial autocorrelation analysis and population genetics models. Heredity 83:145–154
Hardy OJ, Vekemans X (2002) SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes 2:618–620
Hardy OJ, Maggia L, Bandou E, Breyne P, Caron H, Chevallier ME, Doligez A, Dutech C, Kremer A, Latouche-Halle C, Troispoux V, Veron V, Degen B (2006) Fine-scale genetic structure and gene dispersal influences in 10 neotropical tree species. Mol Ecol 15:559–571
Jankowska-Wroblewska S, Meyza K, Sztupecka E, Kubera L, Burczyk J (2016) Clonal structure and high genetic diversity at peripheral populations of Sorbus torminalis (L.) Crantz. iForest (early view). doi:10.3832/ifor1885-009
Jump AS, Rico L, Coll M, Penuelas J (2012) Wide variation in spatial genetic structure between natural populations of the European beech (Fagus sylvatica) and its implications for SGS comparability. Heredity 108:633–639
King R, Zalucki JM (2012) Potential inbreeding in a small population of a mass flowering species, Xanthorrhoea johnsonii (Xanthorrhoeaceae): is your mother my father? Am J Plant Sci 3:303–312. doi:10.4236/ajps.2012.33036
Lemenih M (2005) Production and marketing of gums and gum resins in Ethiopia. In: Chikamai B and Casadei E (eds) Production and marketing of gum resins: frankincense, myrrh and opoponax. Network for Natural Gums and Resins in Africa (NGARA). Publication Series No. 5. NGARA, KEFRI, Nairobi, pp 55–70
Lemes MR, Gribel R, Proctor J, Grattapaglia D (2003) Population genetic structure of mahogany (Swietenia macrophylla King, Meliaceae) across the Brazilian Amazon, based on variation at microsatellite loci: implications for conservation. Mol Ecol 12:2875–2883. doi:10.1046/j.1365-294X.2003.01950.x
Moran EV, Clark JS (2011) Estimating seed and pollen movement in a monoecious plant: a hierarchical Bayesian approach integrating genetic and ecological data. Mol Ecol 20:1248–1262
Moran EV, Clark JS (2012) Between-site differences in the scale of dispersal and gene flow in red oak. PLoS One 7:e36492
Ogbazghi W, Rijkers T, Wessel M, Bongers F (2006) Distribution of the frankincense tree Boswellia papyrifera in Eritrea: the role of environment and land use. J Biogeogr 33:524–535
Omondi SF, Kireger E, Dangasuk OG, Chikamai B, Odee DW, Cavers S, Khasa DP (2010) Genetic diversity and population structure of Acacia senegal (L) Willd. in Kenya. Tropical Plant Biol 3:59–70. doi:10.1007/s12042-009-9037-2
Peakall R, Ruibal M, Lindenmayer DB (2003) Spatial autocorrelation analysis offers new insights into gene flow in the Australian bush rat, Rattus fuscipes. Evolution 57:1182–1195
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research – an update. Bioinformatics 28:2537–2539
Raju AJS, Lakshmi PVK, Ramana V, Chandra PH (2012) Entomophily, ornithophily and anemochory in the self-incompatible Boswellia ovalifoliolata Bal. & Henry (Burseraceae), an endemic and endangered medicinally important tree species. Journal of Threatened Taxa 4(7):2673–2684
Rijkers T, Ogbazghi W, Wessel M, Bongers F (2006) The effect of tapping for frankincense on sexual reproduction in Boswellia papyrifera. J Appl Ecol 43:1188–1195
Schroeder JW, Tran HT, Dick CW (2014) Fine scale spatial genetic structure in Pouteria reticulata (Engl.) Eyma (Sapotaceae), a dioecious, vertebrate dispersed tropical rain forest tree species. Glob Ecol Conserv 1:43–49
Segelbacher G, Cushman SA, Epperson BK, Fortin M-J, Francois O, Hardy OJ, Holderegger R, Taberlet P, Waits LP, Manel S (2010) Applications of landscape genetics in conservation biology: concepts and challenges. Conserv Genet 11:375–385. doi:10.1007/s10592-009-0044-5
Smouse P, Peakall R, Gonzales E (2008) A heterogeneity test for fine-scale genetic structure. Mol Ecol 17:3389–3400. doi:10.1111/j.1365-294X.2008.03839
Smulders MJM, Esselink GD, Everaert I, De Riek J, Vosman B (2010) Characterisation of sugar beet (Beta vulgaris L. ssp. vulgaris) varieties using microsatellite markers. BMC Genet 11:41. doi:10.1186/1471-2156-11-41
Sunnichan VG, Morhanram HY, Shivanna KR (2005) Reproductive biology of Boswellia serrata, the source of salaiguggul, an important gum-resin. Bot J Linn Soc 147:73–82
Teshome M (2013) Structure and composition of woody plants in Boswellia dominated woodland of Western Ethiopia. MSc Thesis. Wageningen University, the Netherlands.
Tiep H Van (2015) Forest fragmentation in Vietnam: effects on tree diversity, populations and genetics. PhD thesis, University of Utrecht, the Netherlands. ISBN 978–90–393-6351-5
Tolera M, Sass-Klaassen U, Eshete A, Bongers F, Sterck FJ (2013) Frankincense tree recruitment failed over the past half century. For Ecol Managem 304:65–72. doi:10.1016/j.foreco.2013.04.036
Vollesen K (1989) Burseraceae. In: Hedberg I, Edwards S (eds) Flora of Ethiopia, volume 3. National Herbarium, Addis Ababa University, Addis Ababa and Uppsala University, Uppsala, pp. 442–478
Vekemans X, Hardy OJ (2004) New insights from fine-scale spatial genetic structure analyses in plant populations. Mol Ecol 13:921–935
Vieira FA, Fajardo CG, de Souza AM, de Carvalho D (2010) Landscape-level and fine-scale genetic structure of the neotropical tree Protium spruceanum (Burseraceae). Int J For Res 2010:120979. doi:10.1155/2010/120979
Acknowledgments
This study was financially supported by Netherlands Fellowship program (NUFFIC), the Netherlands. The field research was partly supported by Abdou-Salam Ouédraogo Fellowship through Bioversity International (Rome, Italy), the CGIAR Research Programme on Forests, Trees and Agroforestry and by CGIAR Fund Donors <http://www.cgiar.org/about-us/governing-2010-june-2016/cgiar-fund/fund-donors-2/>.
Authors’ contributions
FB and MJMS conceived the study. ABA sampled the plants. DW and ABA carried out the molecular analyses. ABA and JD analysed the data. ABA, FB, JD and MJMS wrote the paper. All authors have read and approved the submitted manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Data archiving statement
Marker/genotype data available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.26f7v.
Additional information
Communicated by F. Gugerli
Electronic supplementary material
ESM 1
Heterogeneity tests of fine-scale genetic structure between populations and cohorts. t 2 single-class and ω multi-class test criterion. GA: Guba-Arenja adults, GS: Guba-Arenja seedlings, KA: Kurmuk adults, KS: Kurmuk seedlings, GC: Guba-Arenja adult and seedling cohorts combined and KC: Kurmuk adult and seedling cohorts combined; DC: distance classes. (DOCX 25 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Addisalem, A.B., Duminil, J., Wouters, D. et al. Fine-scale spatial genetic structure in the frankincense tree Boswellia papyrifera (Del.) Hochst. and implications for conservation. Tree Genetics & Genomes 12, 86 (2016). https://doi.org/10.1007/s11295-016-1039-2
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-016-1039-2