Toward a global platform for linking soil biodiversity data
- 1Terrestrial Ecology, Netherlands Institute of Ecology (NIOO-KNAW), Wageningen, Netherlands
- 2German Centre for Integrative Biodiversity Research (iDiv) Halle-Jena-Leipzig, Leipzig, Germany
- 3Global Biodiversity Information Facility, Copenhagen, Denmark
- 4Institute of Biology, University of Leipzig, Leipzig, Germany
- 5Joint Research Center—EU, Ispra, Italy
- 6The Gladstone Institutes, University of California, San Francisco, San Francisco, CA, USA
- 7Department of Ecology and Evolutionary Biology, Cooperative Institute for Research in Environmental Sciences, University of Colorado, Boulder, CO, USA
- 8Department of Soil Ecology, Helmholtz Centre for Environmental Research, Halle/Salle, Germany
- 9Department of Biology, University of Western Ontario, London, ON, Canada
- 10Plant Ecology, Institute of Biology, Freie Universität Berlin, Berlin, Germany
- 11Berlin-Brandenburg Institute of Advanced Biodiversity Research, Berlin, Germany
- 12Senckenberg Museum of Natural History, Görlitz, Germany
- 13J.F. Blumenbach Institute for Zoology and Anthropology, Georg August University, Göttingen, Germany
- 14Agriculture and Agri-food Canada, Eastern Corn and Oilseed Research Centre, Ottawa, ON, Canada
- 15Faculty of Life Sciences, The University of Manchester, Manchester, UK
- 16Laboratory of Nematology, Wageningen University, Wageningen, Netherlands
- 17School of Global Environmental Sustainability, Colorado State University, Fort Collins, CO, USA
Soil biodiversity is immense, with an estimated 10–100 million organisms belonging to over 5000 taxa in a handful of soil. In spite of the importance of soil biodiversity for ecosystem functions and services, information on soil species, from taxonomy to biogeographical patterns, is incomplete and there is no infrastructure to connect pre-existing or future data. Here, we propose a global platform to allow for greater access to soil biodiversity information by linking databases and repositories through a single open portal. The proposed platform would for the first time, link data on soil organisms from different global sites and biomes, and will be inclusive of all data types, from molecular sequences to morphology measurements and other supporting information. Access to soil biodiversity species records and information will be instrumental to progressing scientific research and education. Further, as demonstrated by previous biodiversity synthesis efforts, data availability is key for adapting to, and creating mitigation plans in response to global changes. With the rapid influx of soil biodiversity data, now is the time to take the first steps forward in establishing a global soil biodiversity information platform.
Introduction
Soils are increasingly recognized as crucial components of ecosystems and biodiversity (Wardle et al., 2004; Bardgett and Wardle, 2010), and they represent unique compartments of terrestrial ecosystems by comprising components of the atmosphere, biosphere, hydrosphere, and lithosphere. Soil biodiversity supports many terrestrial ecosystem functions (Wall et al., 2012) and delivers important ecosystem services such as food and fiber production, carbon sequestration, and degradation of pollutants (Wardle, 2002; Wall et al., 2010). However, the data and information regarding diversity that lives in soil remains insufficiently cataloged and coordinated, and this limits our ability to fully assess the key role soil biodiversity plays in supporting terrestrial systems and ecosystem services. In contrast to soil systems, greater effort has been put toward cataloging global diversity in marine and other terrestrial systems (Appeltans et al., 2012; Jetz et al., 2012; Canhos et al., 2014; Hudson et al., 2014) and into making these data free and open access (Guralnick et al., 2007; Wieczorek et al., 2012). Global efforts to synthesize biodiversity data have proven highly successful in the transfer of information, have improved our understanding of species ecology and distribution patterns, and allows for better monitoring and response plans to global change effects (Hampton et al., 2013; Dirzo et al., 2014). Given that we are facing unprecedented environmental alterations through climate change, land use change, soil erosion, invasive species, desertification, and pollution, a better understanding of the global distribution and drivers of soil biodiversity is urgently needed to forecast functional changes of terrestrial ecosystems and to develop appropriate management practices. Therefore, here we review the rationale behind and the benefits of bringing together soil biodiversity data and information through a single global data platform.
Although it is known that soils are extraordinarily diverse, the scale of soil biodiversity is not yet fully understood (Wall et al., 2010). Global patterns of soil biodiversity are at most weakly documented (Decaëns, 2010; Tedersoo et al., 2014), and the locations of many soil biodiversity hotspots have not been identified. Part and parcel to the plethora of hyperdiverse taxonomic groups, global patterns of soil biodiversity are thought to differ significantly from what is reported aboveground (Maraun et al., 2007; Decaëns, 2010; Tedersoo et al., 2012; Ramirez et al., 2014). For example, soil microorganisms do not respond to large-scale environmental gradients in the same way as metazoans and belowground biodiversity hotspots do not necessarily mirror aboveground biodiversity patterns (Fierer and Jackson, 2006; Wu et al., 2011) Further, many species residing in soil remain taxonomically, phylogenetically, and functionally undescribed. This is most notable for microorganisms (McDonald et al., 2012) but it is also true for soil fauna (Behan–Pelletier, 1999; Rougerie et al., 2009; Bik et al., 2012). Therefore, categorizing species into discrete taxonomic units represents a challenge for soil biodiversity documentation where many of the species' characteristics and phylogenies are not yet available (Bardgett and van der Putten, 2014).
Regardless of these challenges, soil biodiversity research has dramatically increased over the last three decades, and the scope of soil biodiversity data is immense. Soil biodiversity data types range from classical specimen based collections (Burkhardt et al., 2014) to molecular and genomics samples (Gilbert et al., 2014). In between are a wide spectrum of community-aggregated data (i.e., trophic levels to relative abundances) organism attributes (e.g., abundance, biomass, and traits), and environmental measurements (e.g., georeference coordinates, biome type, soil characteristics, and climatic variables). Like other biodiversity information, soil biodiversity data can be digital and available online, though much data remains “dark”—not digitized or not available (Heidorn, 2008). Whether in a national repository, stored on a personal computer, or found in a museum drawer, the first step in any data synthesis project is to make dark data digitally accessible (Box 1) (Hill et al., 2012). Next is to establish a mechanism to link digitally available data globally (such as an online portal).
Box 1. Digital soil biodiversity information is currently stored in a wide array of databases, warehouses, catalogs, and other repositories, and contains various types of data (see Supplemental Table 1 for a more extensive list of examples).
• Catalogs: Taxonomy lists with descriptions of the organism. May have occurrence data and may contain images, videos or other media. (Example: Encyclopedia of Life)
• Data Warehouse: An information system that links taxonomy (morphology and/or annotated sequences) and ecological information across databases and individual studies. (Example: Edaphobase)
• Public or Private Databases: Species lists for a given study, experiment or location. May include any number of additional measured parameters such as soil environment measurements and climate information. (Example: Earth Microbiome Project)
• Sequence Archives: Nucleotide sequences that provide valuable information on relevant organisms. These can be useful for determining phylogenies and functional characteristics of organisms. May follow standards of Genomic Standards Consortia. [Example: European Nucleotide Archive (ENA), National Center for Biotechnology Information (NCBI)]
Here we present an independent initiative to assess and store information on global soil biodiversity; to link species, environmental, and other data and make data accessible at a global level. Our goal was to propose a system that could be linked to other biodiversity and ecosystem relevant databases, accommodate new and future methods and technologies, be useful to a wide array of end users (from the public to scientists to policy makers), and be free and open access.
Applied Advances
It is now commonplace to concurrently survey soil biodiversity and explore the role these organisms play in ecosystem functions and global sustainability (Wall et al., 2012; Bardgett and van der Putten, 2014). However, we still lack baseline values for soil biodiversity as well as reference values (either abundance ranges or occurrence) that may prove critical in assessing the current status of soils and implementing management and policy efforts to keeping soils and soil biodiversity in a so-called “normal operating range” (Jackson et al., 2007; Koch et al., 2013). This will be particularly important as we continue to understand the impact of certain global changes on soil biodiversity and their interactions within functioning food webs (Blankinship et al., 2011; García-Palacios et al., 2015). For example, agricultural intensification reduces the abundance of soil fungi relative to bacteria, reduces earthworms, mycorrhizal fungi, and increases the numbers of plant parasitic nematodes (Tsiafouli et al., 2015). Less is known on effects of incipient changes, or changes that encompass temporally complex and indirect feedback effects, such as consequences of global warming, biological invasions, or habitat fragmentation (Blankinship et al., 2011; Lindo et al., 2012; Dickie et al., 2014).
Reference values can be an important tool for determining the success of ecosystem restoration and comparing data across time scales (Frouz et al., 2004; Kardol and Wardle, 2010) and for detecting subtle trends in temporal soil biodiversity assessments (Bardgett, 2005). Specific indicators that can be accessed from a global platform, such as disease-suppression (Mendes et al., 2011) and nutrient retention capacity of soil (De Vries et al., 2013), can also be used by land managers in order to calibrate and further improve sustainability of production methods, or used to develop rapid and economic soil biodiversity assessment tools for use by policy makers and end users (Wall et al., 2012; Bone et al., 2014). As demonstrated by the Global Biodiversity Information Facility (GBIF) and other global data synthesis efforts (Otegui et al., 2013), access and availability of data has helped to predict the impact of climate change (Warren et al., 2013), monitor invasive species (Gatto et al., 2013) and inform on issues like human health (Daszak et al., 2013) and food and farming (Vincent et al., 2013). Further, the efforts by GBIF and Map of Life (MOL) support the work of the CBD, IPBES, GEO-BON, and many others (see GBIF.org). The inclusion of soil biodiversity data in such global assessments is a highly important and necessary next step.
Theoretical and Research Advances
The prospect of accessing global soil biodiversity information through a single portal will create novel opportunities to develop, refine, and test underlying ecological theory. The synthesis of biodiversity data across larger spatial scales and greater taxonomic breadth may uncover emergent properties that cannot currently be foreseen (Brose et al., 2012) and will give better insight into species' ecological preferences and geographical ranges (Brose et al., 2004; Fierer et al., 2013; Tedersoo et al., 2014). Here we identify five topic areas that, while not exhaustive, will be enhanced by a global data platform effort:
(1) Macroecology and biogeographical patterns: Characterizing global patterns is of paramount importance for conservation of soil biodiversity and global change scenarios on the functioning of soil systems in a future world. A comprehensive view of biogeographic patterns will be critical to reveal important scientific questions, to discover where and why there are hot spots of biodiversity, to identify the drivers of belowground diversity, and will ultimately boost the use of macroecological approaches in soil ecology research (Fierer et al., 2013; Tedersoo et al., 2014).
(2) Biodiversity maintenance and loss: A synthesis of soil biodiversity data will help identify drivers and mechanisms underlying both the maintenance and loss of biodiversity in soil and dependent terrestrial systems. The support that belowground diversity gives to aboveground diversity is drastically underestimated, and by overlaying belowground and aboveground biodiversity patterns we can better assess the impact of biodiversity losses. Further, these efforts may prove especially important in terms of invasion ecology, identifying which groups are prone to invade (e.g., earthworms, Hendrix et al., 2008), and the mechanisms facilitating invasion (e.g., Dickie et al., 2014) and prevention efforts.
(3) Ecosystem functions and services: Soil organisms co-determine a plethora of provisioning and regulating ecosystem services (Wardle et al., 2004; Lavelle et al., 2006), but the appreciation of their functional significance remains deficient due to their cryptic nature and overlapping functions (Setälä et al., 2005). While conventional anthropogenic land management practices often have aimed to optimize certain (single) ecosystem functions or services (Cardinale et al., 2012), soil biodiversity exemplifies the value of multifunctional ecosystems (Setälä et al., 2014; Wagg et al., 2014). Recent evidence shows that the structure and composition of the soil community and the presence of specific functional groups, is key to delivering a range of ecosystem services, such as N retention and C storage (De Vries et al., 2013; Lange et al., 2015).
(4) Community ecology: Soil communities are notoriously complex and conventional community ecological theory may be challenged by the spatially complex habitat soil organisms live in (Ettema and Wardle, 2002). Multitrophic soil biodiversity assessment may help to refine existing soil food web models (Digel et al., 2014). Further, global-scale information on the co-occurrence of different taxa in soil will shed light on the relative significance of trophic vs. non-trophic interactions in soil, top-down vs. bottom up forces and their interplays (Moore et al., 2004) and ecological network perspectives may provide useful tools to clarify interactions among the different soil functional groups and to certain ecosystem functions (Barberán et al., 2011; Morriën and van der Putten, 2013).
(5) Aboveground-belowground interactions: As our knowledge of belowground communities increases, so too does our awareness of the important, complex interactions between soil organisms and aboveground biodiversity (Hooper et al., 2000). By revealing belowground biodiversity patterns, we can gain better insight into the linkages between above- and below-ground systems. Plus, soil biodiversity data will be made more valuable if it can be clearly linked to with data pertaining to aboveground communities (such as through the MOL or GBIF).
A Proposed Framework
Our ability to address a range of applied and theoretical questions, or to assess biogeographical patterns, is to a large extent limited by access and integration of the available data. Currently, there is no single repository or platform that allows access to soil biodiversity information, across all species, or at a global scale. Therefore, we propose a framework to initiate linking different databases and repositories via the internet (Figure 1). The end platform will be both a database and a free, open access portal to link various national and local data sources around the world. Linking data from existing databases is not trivial, nor is it a new challenge (Jetz et al., 2012). Previous efforts such as GBIF and MOL have demonstrated that because there are no required guidelines or consistency between studies or pre-established databases, minimum standards, and classifications must be identified. Soil biodiversity standards must then be harmonized with the global standards already in place (e.g., Yilmaz et al., 2011; Wieczorek et al., 2012). While applying even simple standards will lead to the omission of some studies and data, quality of the data will be valued over quantity, ultimately resulting in a higher quality synthesis.
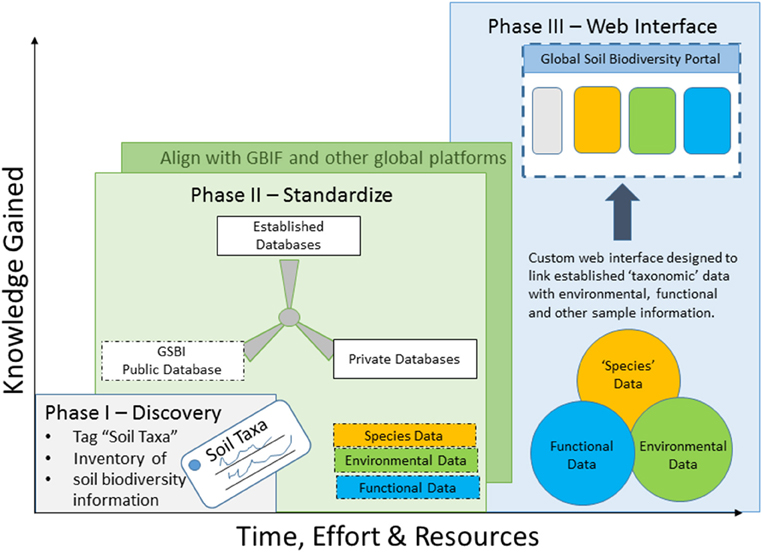
Figure 1. Integration and access to soil biodiversity data will be accomplished in three phases: (I) discovery, (II) standardization, and (III) a final user interface, and the timing of these phases will be directly related to the effort and resources put in to the framework.
Integration and access to soil biodiversity data will be accomplished in three phases: discovery, standardization, and a final user interface:
Phase I—“Discover” where soil biodiversity data is housed: This phase will be two-fold; first to establish a taxonomy list—a list of organisms living in the soil, and second to inventory soil biodiversity information. The taxonomy list will be shared with the GBIF to tag pre-existing soil related biological observations that can thereon be searched and queried [much like the Global Mountain Biodiversity Assessment (GMBA) (gmba.unibas.ch)] and allow for easier integration of new data. The “taxonomy list” and an inventory of soil biodiversity information will be made available through the Global Soil Biodiversity Initiative (GSBI). It is in this stage that data quality will be also assessed, a complicated issue all biodiversity data studies must deal with. We propose to follow guidelines set forth and established by GBIF.
Phase II—Establish a standardization framework by which to link past, present, and future data: Besides taxonomic synonyms it also will be necessary to develop and implement thesauri for the various information fields (i.e., regarding habitat or climate parameters, methods etc.). Standardized ontologies are necessary to link between different data sources and into GBIF (Supplemental Table 1) and other global data centers (such as MOL, ISRIC, EOL, Genebank, and others). Furthermore, to allow data comparability from the individual data sources, standardization of numeric (abundances, pH values, etc.), and nominal (i.e., habitat types, soil types) data will be crucial. Concurrently, we must also establish the minimum set of parameters needed, and formalize data copyright privacy and licensing rules. Together these efforts will provide the critical foundation and quality criteria on which to build the platform.
Short read sequence data: In the case of microbial marker gene sequence data (either 16S, 18S, ITS or similar) it is difficult to extract taxonomic information for a number of reasons (otu picking methods, chimeras, read length, Orgiazzi et al., 2014). Plus due to the enormous amount of sequence data, reprocessing the full datasets would not be tractable. Therefore, we propose to link short read sequence data by location, rather than by taxon identification. This is based on the fact that there is currently no consensus on the correct protocol for handling these data, and integrating processed sequence data would introduce substantial methodological artifacts (Caporaso et al., 2010). Instead, our approach allows convenient access to these data linked to geography and allows users to process the data of interest using a consistent protocol based on individual research questions.
Phase III—Establish a user-friendly interface that allows for the integration and comparison of soil biodiversity data—here called “Soil Portal”: The portal will be designed specifically for manipulation and analyses of the data in order to address the theoretical questions outlined above and to provide stakeholders with the type of information needed for management and policy decisions. It is in this phase that we would finally be able to combine collection data across taxonomic groups, spatial scales and research experiments. As demonstrated previously (Hill et al., 2012), users are reluctant to use any interface that costs time, therefore, we propose a platform that would offer researchers a set of tools, rewards for contributing their data to the community- such as data analyses tools, DOIs for data publication, and a link to other initiatives and data portals.
Outlook
In order to progress this project, first, buy-in from the community of soil biologists is required; our goal is to galvanize and guide soil ecologists to make their data available. Researchers can continue to upload data from their home repositories, data will not have to be uploaded more than once, and there is no need to support a single, comprehensive database—a monetarily expensive and time consuming task. The framework is designed so that participation in the effort to liberate individual datasets will only require minor changes to how researchers work (i.e., time for data input and training for students and young scientists), but has the potential for great individual rewards such as more publications (e.g., “data papers”), increased exposure leading to invitations and collaborations, as well as reciprocal access to a wealth of data from colleagues. Admittedly, in addition to the technical challenges outlined in the introduction, the main limiting factor of this proposal will be resources. Specifically, time and funds must be invested upfront to move this effort forward in an efficient way.
Conclusion
In response to unprecedented global environmental changes and the drastic impacts on biodiversity (Sala et al., 2000), there is a sense of urgency to bring together global biodiversity information that will provide the basis to determine the species and communities that are particularly vulnerable to change and extinctions (Scholes et al., 2008; Cardinale et al., 2012; Jetz et al., 2012) and focus conservation and management practices (Turner et al., 2015). The organisms that live in the soil are no exceptions. The focus of the outlined framework goes beyond species information, and therefore a major challenge and goal will be to integrate the different information types whereby a range of ecological questions can be addressed. Soil biodiversity information is of broad interest to other disciplines, including plant ecologists, agriculturalists, invertebrate ecologists, carbon and climate modelers, and would open new unique opportunities for collaboration between the groups. As such, we have designed a framework that will interface with other disciplines through GBIF and the like. In addition to data access and standardization, a priority of this effort will be analytical and visualization tools for end users. Beyond progressing scientific research these tools should help to communicate results and bring the interest of a larger, more general audience. Altogether, access to rapidly accumulating soil biodiversity information across the globe has the potential to improve research and elevate soil ecology to be on par with our understanding of aboveground systems.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Marten Winter for his contribution to the ideas presented here. This was a joint effort of the Global Soil Biodiversity Initiative (GSBI) and working group sOILDIV, and was kindly supported by sDiv, the Synthesis Centre for Biodiversity Sciences of the German Centre for Integrative Biodiversity Research (iDiv) Halle-Jena-Leipzig (DFG FZT 118). This work was also supported by an ERC grant [ERC-Adv 260-55290 (SPECIALS)] to WP.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/article/10.3389/fevo.2015.00091
References
Appeltans, W., Ahyong, S. T., Anderson, G., Angel, M. V., Artois, T., Bailly, N., et al. (2012). The magnitude of global marine species diversity. Curr. Biol. 22, 2189–2202. doi: 10.1016/j.cub.2012.09.036
Barberán, A., Bates, S. T., Casamayor, E. O., and Fierer, N. (2011). Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J. 6, 343–351. doi: 10.1038/ismej.2011.119
Bardgett, R. D. (2005). The Biology of Soil: A Community and Ecosystem Approach. Oxford: Oxford University Press. doi: 10.1093/acprof:oso/9780198525035.001.0001
Bardgett, R. D., and van der Putten, W. H. (2014). Belowground biodiversity and ecosystem functioning. Nature 515, 505–511. doi: 10.1038/nature13855
Bardgett, R. D., and Wardle, D. A. (2010). Aboveground–belowground Linkages: Biotic Interactions, Ecosystem Processes, and Global Change. New York, NY: Oxford University Press.
Behan–Pelletier, V. M. (1999). Oribatid mite biodiversity in agroecosystems: role for bioindication. Agric. Ecosys. Environ. 74, 411–423. doi: 10.1016/S0167-8809(99)00046-8
Bik, H. M., Porazinska, D. L., Creer, S., Caporaso, J. G., Knight, R., and Thomas, W. K. (2012). Sequencing our way towards understanding global eukaryotic biodiversity. Trends Ecol. Evol. 27, 233–243. doi: 10.1016/j.tree.2011.11.010
Blankinship, J. C., Niklaus, P. A., and Hungate, B. A. (2011). A meta-analysis of responses of soil biota to global change. Oecologia 165, 553–565. doi: 10.1007/s00442-011-1909-0
Bone, J., Barraclough, D., Eggleton, P., Head, M., Jones, D. T., and Voulvoulis, N. (2014). Prioritising soil quality assessment through the screening of sites: the use of publicly collected data. Land Degrad. Dev. 25, 251–266. doi: 10.1002/ldr.2138
Brose, U., Dunne, J. A., Montoya, J. M., Petchey, O. L., Schneider, F. D., and Jacob, U. (2012). Climate change in size-structured ecosystems. Philos. Trans. R. Soc. B Biol. Sci. 367, 2903–2912. doi: 10.1098/rstb.2012.0232
Brose, U., Ostling, A., Harrison, K., and Martinez, N. D. (2004). Unified spatial scaling of species and their trophic interactions. Nature 428, 167–171. doi: 10.1038/nature02297
Burkhardt, U., Russell, D. J., Decker, P., Döhler, M., Höfer, H., Lesch, S., et al. (2014). The Edaphobase project of GBIF–Germany—a new online soil–zoological data warehouse. Appl. Soil Ecol. 83, 3–12. doi: 10.1016/j.apsoil.2014.03.021
Canhos, V. P., Souza, S., Giovanni, R., and Canhos, D. A. L. (2014). Global Biodiversity Informatics: setting the scene for a “new world” of ecological forecasting. Biodivers. Inform. 1, 1–13. doi: 10.17161/bi.v1i0.3
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Lozupone, C. A., Turnbaugh, P. J., et al. (2010). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U.S.A. 108(Suppl. 1), 4516–4522. doi: 10.1073/pnas.1000080107
Cardinale, B. J., Duffy, J. E., Gonzalez, A., Hooper, D. U., Perrings, C., Venail, P., et al. (2012). Biodiversity loss and its impact on humanity. Nature 486, 59–67. doi: 10.1038/nature11148
Daszak, P., Zambrana-Torrelio, C., Bogich, T. L., Fernandez, M., Epstein, J. H., Murray, K. A., et al. (2013). Interdisciplinary approaches to understanding disease emergence: the past, present, and future drivers of Nipah virus emergence. Proc. Natl. Acad. Sci.U.S.A. 110, 3681–3688. doi: 10.1073/pnas.1201243109
Decaëns, T. (2010). Macroecological patterns in soil communities. Glob. Ecol. Biogeogr. 19, 287–302. doi: 10.1111/j.1466-8238.2009.00517.x
de Vries, F. T., Thébault, E., Liiri, M., Birkhofer, K., Tsiafouli, M. A., Bjørnlund, L., et al. (2013). Soil food web properties explain ecosystem services across European land use systems. Proc. Natl. Acad. Sci. U.S.A. 110, 14296–14301. doi: 10.1073/pnas.1305198110
Dickie, I. A., St John, M. G., Yeates, G. W., Morse, C. W., Bonner, K. I., Orwin, K., et al. (2014). Belowground legacies of Pinus contorta invasion and removal result in multiple mechanisms of invasional meltdown. AoB Plants 6:plu056. doi: 10.1093/aobpla/plu056
Digel, C., Curtsdotter, A., Riede, J., Klarner, B., and Brose, U. (2014). Unravelling the complex structure of forest soil food webs: higher omnivory and more trophic levels. Oikos 123, 1157–1172. doi: 10.1111/oik.00865
Dirzo, R., Young, H. S., Galetti, M., Ceballos, G., Isaac, N. J. B., and Collen, B. (2014). Defaunation in the Anthropocene. Science 345, 401–406. doi: 10.1126/science.1251817
Ettema, C. H., and Wardle, D. A. (2002). Spatial soil ecology. Trends Ecol. Evol. 17, 177–183. doi: 10.1016/S0169-5347(02)02496-5
Fierer, N., and Jackson, R. B. (2006). The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. U.S.A. 103, 626–631. doi: 10.1073/pnas.0507535103
Fierer, N., Ladau, J., Clemente, J. C., Leff, J., Owens, S. M., Pollard, K. S., et al. (2013). Reconstructing the microbial diversity and function of pre–agricultural tallgrass prairie soils in the United States. Science 342, 621–624. doi: 10.1126/science.1243768
Frouz, J., Ali, A., Frouzova, J., and Lobinske, R. J. (2004). Horizontal and vertical distribution of soil macroarthropods along a spatio–temporal moisture gradient in subtropical Central Florida. Environ. Entomol. 33, 1282–1295. doi: 10.1603/0046-225X-33.5.1282
García-Palacios, P., Vandegehuchte, M. L., Shaw, E. A., Dam, M., Post, K. H., Ramirez, K. S., et al. (2015). Are there links between responses of soil microbes and ecosystem functioning to elevated CO2, N deposition and warming? A global perspective. Glob. Change Biol. 21, 1590–1600. doi: 10.1111/gcb.12788
Gatto, F., Katsanevakis, S., Vandekerkhove, J., Zenetos, A., and Cardoso, A. C. (2013). Evaluation of online information sources on alien species in Europe – the need of harmonization and integration. Environ. Manag. 51, 1137–1146. doi: 10.1007/s00267-013-0042-8
Gilbert, J. A., Jansson, J. K., and Knight, R. (2014). The earth microbiome project: successes and aspirations. BMC Biol. 12:69. doi: 10.1186/s12915-014-0069-1
Guralnick, R. P., Hill, A. W., and Lane, M. (2007). Toward a collaborative, global infrastructure for biodiversity assessment. Ecol. Lett. 10, 663–672. doi: 10.1111/j.1461-0248.2007.01063.x
Hampton, S. E., Strasser, C. A., Tewksbury, J. J., Gram, W. K., Budden, A. E., Batcheller, A. L., et al. (2013). Big data and the future of ecology. Front. Ecol. Environ. 11, 156–162. doi: 10.1890/120103
Heidorn, P. B. (2008). Shedding light on the dark data in the long tail of science. Libr. Trends 57, 280–299. doi: 10.1353/lib.0.0036
Hendrix, P. F., Callaham, M. A. Jr., Drake, J. M., Huang, C.–Y., James, S. W., Snyder, B. A., et al. (2008). Pandora's box contained bait: the global problem of introduced earthworms. Annu. Rev. Ecol. Evol. Syst. 39, 593–613. doi: 10.1146/annurev.ecolsys.39.110707.173426
Hill, A., Guralnick, R., Smith, A., Sallans, A., Gillespie, R., Denslow, M., et al. (2012). The notes from nature tool for unlocking biodiversity records from museum records through citizen science. Zookeys 209, 219–233. doi: 10.3897/zookeys.209.3472
Hooper, D. U., Bignell, D. E., Brown, V. K., Brussard, L., Dangerfield, J. M., Wall, D. H., et al. (2000). Interactions between aboveground and belowground biodiversity in terrestrial ecosystems: patterns, mechanisms, and feedbacks. Bioscience 50, 1049–1061. doi: 10.1641/0006-3568(2000)050[1049:IBAABB]2.0.CO;2
Hudson, L. N., Newbold, T., Contu, S., Hill, S. L. L., Lysenko, I., De Palma, A., et al. (2014). The PREDICTS database: a global database of how local terrestrial biodiversity responds to human impacts. Ecol. Evol. 4, 4701–4735. doi: 10.1002/ece3.1303
Jackson, L. E., Pascual, U., and Hodgkin, T. (2007). Utilizing and conserving agrobiodiversity in agricultural landscapes. Agric. Ecosyst. Environ. 121, 196–210. doi: 10.1016/j.agee.2006.12.017
Jetz, W., McPherson, J. M., and Guralnick, R. P. (2012). Integrating biodiversity distribution knowledge: toward a global map of life. Trends Ecol. Evol. 27, 151–159. doi: 10.1016/j.tree.2011.09.007
Kardol, P., and Wardle, D. A. (2010). How understanding aboveground–belowground linkages can assist restoration ecology. Trends Ecol. Evol. 25, 670–679. doi: 10.1016/j.tree.2010.09.001
Koch, A., McBratney, A., Adams, M., Field, D., Hill, R., Crawford, J., et al. (2013). Soil security: solving the global soil crisis. Glob. Policy 4, 434–441. doi: 10.1111/1758-5899.12096
Lange, M., Eisenhauer, N., Sierra, C. A., Bessler, H., Engels, C., Griffiths, R. I., et al. (2015). Plant diversity increases soil microbial activity and soil carbon storage. Nat. Commun. 6:6707. doi: 10.1038/ncomms7707
Lavelle, P., Decaëns, T., Aubert, M., Barot, S., Blouin, M., Bureau, F., et al. (2006). Soil invertebrates and ecosystem services. Eur. J. Soil Biol. 42, 3–15. doi: 10.1016/j.ejsobi.2006.10.002
Lindo, Z., Whiteley, J. A., and Gonzalez, A. (2012). Traits explain community disassembly and trophic contraction following experimental environmental change. Glob. Change Biol. 18, 2448–2457. doi: 10.1111/j.1365-2486.2012.02725.x
Maraun, M., Schatz, H., and Scheu, S. (2007). Awesome or ordinary? Global diversity patterns of oribatid mites. Ecography 30, 209–216. doi: 10.1111/j.0906-7590.2007.04994.x
McDonald, D., Price, M. N., Goodrich, J., Nawrocki, E. P., DeSantis, T. Z., Probst, A., et al. (2012). An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 6, 610–618. doi: 10.1038/ismej.2011.139
Mendes, R., Kruijt, M., De Bruijn, I., Dekkers, E., Van der Voort, M., Schneider, J. H. M., et al. (2011). Deciphering the rhizosphere microbiome for disease–suppressive bacteria. Science 332, 1097–1100. doi: 10.1126/science.1203980
Moore, J. C., Berlow, E. L., Coleman, D. C., de Ruiter, P. C., Dong, Q., Hastings, A., et al. (2004). Detritus, trophic dynamics, and biodiversity. Ecol. Lett. 7, 584–600. doi: 10.1111/j.1461-0248.2004.00606.x
Morriën, E., and van der Putten, W. H. (2013). Soil microbial community structure of range-expanding plant species differs from co-occurring natives. J. Ecol. 101, 1093–1102. doi: 10.1111/1365-2745.12117
Orgiazzi, A., Dunbara, M. B., Panagosa, P., de Groot, A. G., and Lemanceau, P. (2014). Soil biodiversity and DNA barcodes: opportunities and challenges. Soil Biol. Biochem. 80, 244–250. doi: 10.1016/j.soilbio.2014.10.014
Otegui, J., Ariño, A. H., Encinas, M. A., and Pando, F. (2013). Assessing the primary data hosted by the Spanish node of the Global Biodiversity Information Facility (GBIF). PloS ONE 8:e55144. doi: 10.1371/journal.pone.0055144
Ramirez, K. S., Leff, J. W., Barberán, A., Bates, S. T., Betley, J., Crowther, T. W., et al. (2014). Biogeographic patterns in belowground diversity in New York City's Central Park are similar to those observed globally. Proc. R. Soc. B. 281:20141988. doi: 10.1098/rspb.2014.1988
Rougerie, R., Decaëns, T., Deharveng, L., James, S., Porco, D., Chang, C. H., et al. (2009). DNA barcodes for soil animal taxonomy. Braz. J. Agric. Res. 44, 789–801. doi: 10.1590/S0100-204X2009000800002
Sala, O. E., Chapin, F. S. III, Armesto, J. J., Berlow, E., Bloomfield, J., Dirzo, R., et al. (2000). Global biodiversity scenarios for the year 2100. Science 287, 1770–1774. doi: 10.1126/science.287.5459.1770
Scholes, R. J., Mace, G. M., Turner, W., Geller, G. N., Jürgens, N., Larigauderie, A., et al. (2008). Toward a global biodiversity observing system. Science 321, 1044–1045. doi: 10.1126/science.1162055
Setälä, H., Bardgett, R. D., Birkhofer, K., Brady, M., Byrne, L., de Ruiter, P. C., et al. (2014). Urban and agricultural soils: conflicts and trade–offs in the optimization of ecosystem services. Urban Ecosyst. 17, 239–253. doi: 10.1007/s11252-013-0311-6
Setälä, H., Berg, M. B., and Jones, T. H. (2005). “Trophic structure and functional redundancy in soil communities,” in Biological Diversity and Function in Soils, eds R. Bardgett, M. Usher, and D. Hopkins (Cambridge: Cambridge University Press), 236–249. doi: 10.1017/CBO9780511541926.014
Tedersoo, L., Bahram, M., Põlme, S., Kõljalg, U., Yorou, N. S., et al. (2014). Global diversity and geography of soil fungi. Science 346:1256688. doi: 10.1126/science.1256688
Tedersoo, L., Bahram, M., Toots, M., Diédhiou, A. G., Henkel, T. W., Kjøller, R., et al. (2012). Towards global patterns in the diversity and community structure of ectomycorrhizal fungi. Mol. Ecol. 21, 4160–4170. doi: 10.1111/j.1365-294X.2012.05602.x
Tsiafouli, M. A., Thebault, E., Sgardelis, S. P., de Ruiter, P. C., van der Putten, W. H., Birkhofer, K., et al. (2015). Intensive agriculture reduces soil biodiversity across Europe. Glob. Change Biol. 21, 973–985. doi: 10.1111/gcb.12752
Turner, W., Rondinini, C., Pettorelli, N., Mora, B., Leidner, A. K., Szantoi, Z., et al. (2015). Free and open-access satellite data are key to biodiversity conservation. Biol. Conserv. 182, 173–176. doi: 10.1016/j.biocon.2014.11.048
Vincent, H., Wiersema, J., Kell, S., Fielder, H., Dobbie, S., Castañeda-Álvarez, N. P., et al. (2013). A prioritized crop wild relative inventory to help underpin global food security. Biol. Conserv. 167, 265–275. doi: 10.1016/j.biocon.2013.08.011
Wagg, C., Bender, S. F., Widmer, F., and van der Heijden, M. G. A. (2014). Soil biodiversity and soil community composition determine ecosystem multifunctionality. Proc. Natl. Acad. Sci. U.S.A. 111, 5266–5270. doi: 10.1073/pnas.1320054111
Wall, D. H., Bardgett, R. D., Behan-Pelletier, V., Herrick, J. E., Jones, H., Ritz, K., et al. (2012). Soil Ecology and Ecosystem Services. Oxford, UK: Oxford University Press. Available online at: http://www.amazon.com/Soil-Ecology-Ecosystem-Services-Diana/dp/0199575924 (Accessed November 5, 2013).
Wall, D. H., Bardgett, R. D., and Kelly, E. (2010). Biodiversity in the dark. Nat. Geosci. 3, 297–298. doi: 10.1038/ngeo860
Wardle, D. A. (2002). Communities and Ecosystems: Linking the Aboveground and Belowground Components. Princeton, NJ: Princeton University Press.
Wardle, D. A., Bardgett, R. D., Klironomos, J. N., Setälä, H., Van Der Putten, W. H., and Wall, D. H. (2004). Ecological linkages between aboveground and belowground biota. Science 304, 1629–1633. doi: 10.1126/science.1094875
Warren, R., VanDerWal, J., Price, J., Welbergen, J. A., Atkinson, I., Ramirez–Villegas, J., et al. (2013). Quantifying the benefit of early climate change mitigation in avoiding biodiversity loss. Nat. Clim. Change 3, 678–682. doi: 10.1038/nclimate1887
Wieczorek, J., Bloom, D., Guralnick, R., Blum, S., Döring, M., Giovanni, R., et al. (2012). Darwin Core: an evolving community–developed biodiversity data standard. PLoS ONE 7:e29715. doi: 10.1371/journal.pone.0029715
Wu, T., Ayres, E., Bardgett, R. D., Wall, D. H., and Garey, J. R. (2011). Molecular study of worldwide distribution and diversity of soil animals. Proc. Natl. Acad. Sci. U.S.A. 108, 17720–17725. doi: 10.1073/pnas.1103824108
Keywords: soil ecology, belowground biodiversity, biogeography, data synthesis, taxonomy, sequence data, ecological theory
Citation: Ramirez KS, Döring M, Eisenhauer N, Gardi C, Ladau J, Leff JW, Lentendu G, Lindo Z, Rillig MC, Russell D, Scheu S, St. John MG, de Vries FT, Wubet T, van der Putten WH and Wall DH (2015) Toward a global platform for linking soil biodiversity data. Front. Ecol. Evol. 3:91. doi: 10.3389/fevo.2015.00091
Received: 24 May 2015; Accepted: 17 July 2015;
Published: 30 July 2015.
Edited by:
Angela McGaughran, Commonwealth Scientific and Industrial Research Organisation, AustraliaReviewed by:
Katy Morgan, University of New Orleans, USASara Sanchez Moreno, Instituto Nacional de Investigación y Tecnología Agraria y Alimentaria, Spain
Copyright © 2015 Ramirez, Döring, Eisenhauer, Gardi, Ladau, Leff, Lentendu, Lindo, Rillig, Russell, Scheu, St. John, de Vries, Wubet, van der Putten and Wall. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kelly S. Ramirez, Terrestrial Ecology, Netherlands Institute of Ecology, Droevendaalsesteeg 10, 6708 PB Wageningen, Netherlands, k.ramirez@nioo.knaw.nl
 Kelly S. Ramirez
Kelly S. Ramirez Markus Döring3
Markus Döring3  Nico Eisenhauer
Nico Eisenhauer Ciro Gardi
Ciro Gardi Jonathan W. Leff
Jonathan W. Leff Guillaume Lentendu
Guillaume Lentendu Zoë Lindo
Zoë Lindo Matthias C. Rillig
Matthias C. Rillig Stefan Scheu
Stefan Scheu Mark G. St. John
Mark G. St. John Franciska T. de Vries
Franciska T. de Vries Tesfaye Wubet
Tesfaye Wubet Wim H. van der Putten
Wim H. van der Putten Diana H. Wall
Diana H. Wall