Abstract
The ex situ conservation of plant genetic resources in gene banks involves the selection of accessions to be conserved and the maintenance of these accessions for current and future users. Decisions concerning both these issues require knowledge about the distribution of genetic diversity within and between accessions sampled from the gene pool, but also about the changes in variation of these samples as a result of regenerations. These issues were studied in an existing gene bank collection of a cross-pollinating crop using a selection of groups of very similar Dutch white cabbage accessions, and additional groups of reference material representing the Dutch, and the global white cabbage gene pool. Six accessions were sampled both before and after a standard regeneration. 30 plants of each of 50 accessions plus 6 regeneration populations included in the study were characterised with AFLPs, using scores for 103 polymorphic bands. It was shown that the genetic changes as a result of standard gene bank regenerations, as measured by AFLPs, are of a comparable magnitude as the differences between some of the more similar accessions. The observed changes are mainly due to highly significant changes in allele frequencies for a few fragments, whereas for the majority of fragments the alleles occur in similar frequencies before and after regeneration. It is argued that, given the changes of accessions over generations, accessions that display similar levels of differentiation may be combined safely.
Similar content being viewed by others
Introduction
Ex situ conservation of plant genetic resources in gene banks involves selection of accessions to be conserved and the maintenance of these accessions for current and future users. The first element, the choice of accessions, used to be taken rather lightly in the past. It was considered easy to collect or buy seeds and store these in a freezer. This resulted in more than six million accessions conserved ex situ worldwide (FAO 1996) of which over 10,000 Brassica oleracea accessions in Europe (Boukema and Hintum 1998). However, when it became apparent that all this material had to be characterised and regenerated to be useful, limiting the number of accessions gradually became a new target in plant genetic resources conservation (e.g. McGregor et al. 2002). Especially for cross-pollinating crops this appeared an important concern, since regenerating cross-pollinating accessions is far more costly in comparison to self-pollinating crops due to precautions that need to be taken to avoid pollen contamination. Besides reducing the number of accessions, alternative options for managing costs are valid, such as structuring the collection and selecting a core collection (Hintum et al. 2000). In general, selecting accessions involves criteria such as the threat of genetic erosion, the potential usefulness in breeding and research, the representation in other collections, but also the genetic diversity in the accession as related to the other accessions.
The second element of ex situ conservation is keeping them available. When germinability starts to decrease, or when seeds run out of stock, accessions need to be regenerated. It is of highest importance that the genetic integrity of the gene bank accessions is maintained. Some change of the genetic composition is generally unavoidable, due to hardly controllable processes such as genetic drift and natural selection. In addition, contamination and unintended human selection occur. Therefore, the dangerous step of regeneration needs to be avoided as much as possible by improving the storage condition to increase lifespan of the stored seeds. Effects of regeneration on the genetic composition of gene bank accessions have been studied previously for self-pollinating species (Börner et al. 2000; Parzies et al. 2000), and cross-pollinating species (Chebotar et al. 2003; Diaz et al. 1997; Reedy et al. 1995). These studies have shown that genetic changes resulting from regeneration can be quite substantial. Diaz et al. (1997) showed considerable genetic shift as a result of regeneration of Brassica accessions at different European institutions, Chebotar et al. (2003) found large changes in allele frequencies and influx of up to 50% of new alleles in the course of 7–13 subsequent regenerations of rye, Parzies et al. (2000) found considerable genetic drift in regenerated barley samples. In the studies of Reedy et al. (1995) and Börner et al. (2000), who studied regenerations of maize and wheat, respectively, the effects of drift and shift were much smaller and certainly not alarming.
Both elements, initial selection and regeneration, contain a clear genetic aspect and are inter-related. Questions such as ‘how different should accessions be to justify inclusion in the collection’ or ‘how similar should they be to be considered a duplicate, or a candidate for bulking’ are related to ‘how much genetic shift and drift is acceptable during regeneration’. In the case of the cross-pollinator B. oleracea it has been shown that bulking of similar accessions can be justifiable (Hintum et al. 1996; Phippen et al. 1997) without losing the genetic or historical identity of the accessions. These studies used the conservative approach that only very similar accessions were combined. However, the question is whether one could go further, taking into consideration the changes over regenerations.
To study these issues in an existing gene bank collection a selection of groups of very similar Dutch white cabbage accessions (‘duplication groups’) was made based on the names and origin of these accessions. Based on historical data, these would be the groups within which bulking would be most feasible. To put this selection into perspective, an additional group representing the entire diversity in Dutch white cabbages, and of white cabbages in the world was made. This resulted in a total of 50 accessions. Six of these accessions were sampled both before and after a regular regeneration, to quantify the effects thereof. All accessions were characterised using AFLP. With the help of the AFLP data the following questions were addressed:
-
1.
What is the magnitude of genetic change occurring during regeneration relative to the diversity in the collection, and does it affect all alleles or just a few?
-
2.
To what extent would bulking be justifiable without changing the genetic identity of the accessions, i.e., how unique are the accessions within their ‘duplication group’, relative to the changes during regeneration, how unique are the ‘duplication groups’ in the context of the total genetic diversity of Dutch cabbages and finally how unique are Dutch white cabbages in a global perspective?
The analysis focussed on differences between accessions in duplication groups related to differences over regenerations and visa versa. The data that were generated also allow an analysis of differences between accessions in duplication groups related to differences in the global or Dutch collection. The latter analysis is not discussed here and will be the topic of a subsequent paper.
Materials and methods
Material
A selection was made of seven groups of very similar Dutch white cabbage accessions (‘duplication groups’). The accessions were expected to be similar based on their names and origin. It concerned the duplication groups ‘Brunswijker’, ‘Gouden Akker’, ‘Roem van Enkhuizen’, ‘Delikatesse’, ‘Amager Kortstronk’, ‘Herfstdeen’ and ‘Langedijker’. All CGN accessions in these groups were characterised, except in the group ‘Langedijker’. This group was too large, and a selection of 14 out of 34 accessions was made from this group. To put these accessions into perspective, an additional group of nine accessions representing the diversity in Dutch white cabbages and another group of nine accessions representing white cabbages in the rest of the world was made by selecting as diverse material as possible, based on our knowledge of the material. This resulted in a total number of 50 accessions. From six of these accessions samples from before and after a regular gene bank regeneration were included (Table 1). The accessions have been referred to by their CGN number. If regenerations are concerned these numbers are followed by an ‘o’ (original) or a ‘n’ (new) indicating the accession before or after regeneration, respectively.
At CGN the following procedures are used for regenerating cabbage accessions: (1) Seeds are sown in seed trays at a temperature of 16–20°C in February. (2) After germination seedlings are transplanted to peat pots in a greenhouse with a temperature of 12–15°C. (3) In spring the plants are transplanted to the field. (4) In the autumn the plants are uprooted from the fields, potted and overwintered in a greenhouse during which period they receive a vernalization treatment at temperatures between 5–10°C. (5) In spring, when flowering shoots start to appear, 80 plants or more are placed in isolation rooms (gauze cages) in a non-heated greenhouse. Blowflies are used for pollination. (6) Accessions are harvested when the majority of plants have ripe seeds.
Each accession was characterised on the basis of at least 30 plants. Reproducibility of AFLP bands was monitored by sampling two plants twice for each population. This resulted in 1,832 samples to be characterised. Additionally, on each gel one sample of a standard genotype (plant 31 of accession CGN07040) was included for reference.
DNA extraction
Young leaves from seedlings were collected in 2 ml Eppendorf tubes, frozen in liquid nitrogen, lyophilized and ground with 5–7 glass pearls in a Retch shaking mill prior to the DNA extraction. DNA extraction was performed according to Fulton et al. (1995).
AFLP protocol
The AFLP method was performed essentially as described by Vos et al. (1995). Primary template DNA was prepared in a one-step restiction-ligation reaction. Total genomic DNA (300 ng) was digested with 5 U EcoRI and 5 U MseI (Life Technologies) in 40 μl 10 mM Tris–HCl buffer, 10 mM magnesium acetate, 50 mM potassium acetate, pH 7.5 for 1 h at 37°C. Subsequently, 10 μl of a ligation mixture containing 5 pmol EcoRI adapter, 50 pmol MseI adapter (Isogen) and 2 U T4 DNA ligase (Life Technologies) in the same solution as before but with 0.4 mM ATP, was added. This restriction/ligation reaction was incubated for 3 h at 37°C. The resulting primary template was diluted to 200 μl with 10 mM Tris–HCl, 0.1 mM EDTA pH 8.0 and stored at −20°C until use.
AFLP fingerprints were made using a two-step PCR amplification. The first step (preamplification) was performed on primary template using a primer pair based on the sequences of the EcoRI and MseI adapter with one additional selective nucleotide at the 3′ end. Amplification reactions (20 μl) contained 5 μl primary template, 30 ng of each primer, 0.4 U Taq polymerase, 0.2 mM of all four dNTPs, 10 mM Tris–HCl pH 8.3, 50 mM KCl, 1.5 mM MgCl2 and 0.001% gelatine. Preamplification products were diluted 20-fold with 10 mM Tris–HCl, 0.1 mM EDTA pH 8.0 and used as template in the second amplification reaction. This second step (selective amplification) was performed with primers having three selective nucleotides each. Amplification conditions were as described above except that only 5 ng of 33P-labeled EcoRI primer was used. Standard cycling conditions were: 1 cycle 94°C for 30 s, 65°C for 30 s and 72°C for 1 min. The 65°C annealing temperature was subsequently reduced by 0.7°C for the next 12 cycles, and continued at 56°C for a remaining 23 cycles.
Reaction products were loaded on a 6% polyacrylamide gel (Sequagel-6, Biozym) in 1 × TBE electrophoresis buffer using a SequiGen 38 × 50 cm gel apparatus (BioRad Laboratories). Gels were dried on Whatmann 3MM paper and X-ray films were exposed for 2–5 days at room temperature.
In order to identify primer combinations that yielded well scorable polymorphisms approximately 100 EcoRI/MseI primer combinations were tested on six samples. Suitable combinations were selected based on the number of unambiguously scorable polymorphic bands. Finally two primer combinations were selected for analysis, E39/M36 and E39/M38.
Using these two primer combinations, 150 bands could be scored consistently (77 for the first, 73 for the second primer combination).
Bands were scored by two persons subsequently. Reproducibility of bands over gels was, apart from running the reference sample, tested by repeating PCR reactions from preamplification reactions as well as from the original DNA samples. These tests showed good reproducibility (data not shown).
Scoring
Out of the 150 bands scored, 36 were monomorphic, 19 for the E39/M36 and 17 for the E39/M38 combination. During the analysis, an additional 11 bands showed not to be reliable for analysis. All these were removed from the data set, resulting in a total of 103 polymorphic bands scored.
Since in each population samples 1 and 2, and samples 3 and 4 were respectively sampled from the same individual, the AFLP profiles could be checked for reliability. 1,1031 comparisons could be made without missing values, of which 17 appeared different in the two plants (0.15%). Plants, number 2 and 4 were removed from the data set, except if the number of missing values was higher in the 1 or 3.
The resulting number of plants included in the analysis was 1,714, scored for 103 bands. Since the percentage of missing values was 4.7%, the resulting number of data points was 16,8302, an average of 3,005 per accession, and 98 per plant.
Statistical analysis
Since AFLP bands are dominant, allele-frequencies were estimated from the band frequencies, by using the frequency (q) of absent bands as an estimate of the squared frequency of the recessive allele, assuming Hardy–Weinburg equilibrium in the population.
Gene diversity (H), calculated as 2q(1 − q), is used as a measure of the genetic diversity in accessions or groups of accessions (for discussions, see Crow 1986). To correct for small sample sizes, an unbiased estimate of H is obtained by multiplying the value by the factor n/(n − 1) where n is the number of individuals measured (Nei 1987). These marker diversities are averaged over all markers to calculate the genetic diversity within a group (H T) or within an accession (H S).
G ST was used as a measure of genetic differentiation between accessions (see Nei 1975 for a discussion). It is the proportion of the total genetic diversity that can be attributed to differences among accessions: (H T − H S)/H T in which H T is the total genetic diversity in a set of accessions and H S is the average diversity within these accessions.
The probability of the realised G ST of two populations (an accession before and after regeneration) occurring by chance was determined by simulating the division in groups using the actual fingerprints of the accessions before and after regeneration, and calculating the G ST of the simulated groups. This was repeated 10,000 times.
The probability of changes of individual allele frequencies was estimated using the chi-square (χ2) analysis of the 2 × 2 contingency table with the number of present and absent alleles before and after regeneration (one degree of freedom). The Bonferroni (1936) correction was considered but not applied given the lack of independence between the tests.
For the clustering, an agglomerative approach was used in which the genetic distance between clusters was calculated using Roger’s distance (Rogers 1972; Wright 1978). This distance in this two-state case is basically the sum of absolute band frequency differences. The clustering algorithm used the cluster size as weight. This implied that after each clustering step the frequencies of bands in the newly formed cluster were determined on the basis of all individuals in that cluster and distances of the new cluster with all others were calculated.
All data analyses were performed using software written in MS Visual Basic, interfacing with MS-Excel.
Results
Diversity within and between accessions and regenerations
The genetic diversity (H S) within the 50 accessions varied from 0.05 in CGN11120, member of the ‘Gouden Akker’ group, to 0.18 in CGN07032 of the ‘Brunswijker’ group. The average genetic diversity within single accessions was 0.13 and the total diversity (H T) was 0.24 (Table 2). The genetic differentiation (G ST) in the complete set was thus 0.45.
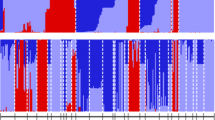
The genetic distances between individual accessions, including the regenerated material, as measured with Rogers’ distance, were the smallest between the populations before and after regeneration, in the range 0.04–0.05. The smallest distance between two different accessions was 0.05 between CGN07039 and CGN14091, both of the ‘Gouden Akker’ group. This implied that the distance between the most similar accessions were of the same order of magnitude as the changes caused by regeneration. The largest distance was 0.35 between CGN11160 of the ‘Delikatesse’ group and CGN07027, an Egyptian landrace. Weighted clustering of accessions and regenerations based on Rogers’ distance resulted in Fig. 1, where the genetic distances between the accessions before and after regeneration are shown, together with the diversity in the most homogeneous duplication group ‘Gouden Akker’ and the most diverse group ‘Langedijker’.
Effects of regeneration
Comparison of the accessions before and after regeneration showed only minor changes in the diversity within the accessions (Table 3). However, due to the differences between the levels of diversity within the accessions, the genetic differentiation (G ST) over regeneration differed considerably, from 0.00 to 0.05. The probability of such differentiation being only due to sampling effects was in two out of the six cases below 5%, so most of the changes were not significant. The changes over regenerations were also examined by comparing the allele frequencies of individual loci. This showed that most of the allele frequencies did not change significantly, but in every population there were a few that did. These changes were highly significant (Fig. 2), between 2 and 21% of the polymorphic bands were significantly different at the 1% level. A Bonferroni correction was considered to avoid the chance of erroneously considering differences significant as a direct result of the high number of tests performed (Bonferroni 1936), but this was not considered appropriate here since the observations could not be assumed to be uncorrelated (Bland and Altman 1995; Perneger 1998). However, even if the Bonferroni correction would have been applied most of the highest chi-squares would still have been significant since the 5% significance level with 56 independent tests (the highest number of polymorphic bands in a regeneration) would correspond to a chi-square of 11.0. A closer examination of all loci with chi-squares over 4.0, i.e., significant at 5%, did not reveal any consistent changes. Only a few alleles had changed significantly in more than one regeneration. One allele increased in frequency in three regenerations significantly, in two other it increased slightly, and in the sixth regeneration the allele was already fixed before regeneration.
Distribution of χ2 values of the frequencies of polymorphic AFLP bands, before and after regeneration (first six histograms) and in the two most similar and the two most distinct accessions. The X-axis presents subsequent intervals of χ2 values (0–1 to >15), the Y-axis the absolute frequency thereof. These values have 1 degree of freedom, and values above 3.84 are significant at the 5% level, and values above 6.64 are significant at the 1% level
Discussion
When studying the effects of regeneration of a cross-pollinating accession, the question whether or not the change by one regeneration cycle is significant depends on the methodology applied. There will be changes, in the best case only small and only due to sampling effects. But even small changes may become significant by characterising many plants and using exact measurements. A more relevant question is what the magnitude of the change occurring during regeneration is, related to the differences between the accessions in the collection. If differences resulting from regeneration exceed those between related accessions, either the improvement of the regeneration protocol or a reduction of the number of accessions should be considered; if a gene bank is not able to maintain the accessions as separate entities over regenerations—because the differences between accessions are too small relative to the changes over regenerations, bulking the most similar accessions should be considered (Sackville Hamilton et al. 2002; Hintum et al. 2002).
Regeneration protocol
The regeneration of white cabbage is a rather complicated procedure (see material section). In this protocol there are a few aspects that will have an influence on the genetic composition of the accession. First there is infra-accession competition, the plants with many flowers will produce more pollen and therefore more seed. Secondly, natural selection will occur, for example, there are seeds that do not germinate and plants that do not survive under these conditions. But also unintentional selection can take place. For example, the seeds are harvested once. Seeds that are not ripe yet will not contribute to the next generation, resulting in a selection for earlyness. The next disturbing factor is genetic drift, although the minimum number of 80 plants per accession should be sufficient to maintain nearly all alleles in the accession. The loss of variation that may be expected after a single regeneration equals 1/2N e, in which N e is the effective population size (Falconer 1981). Assuming that N e = 80, the loss of variation due to random processes is 0.6% per regeneration. Finally, there can be contamination, either of pollen, which is prevented by regenerating in isolation rooms, or of seeds during harvesting, drying, cleaning and packing.
Changes over regenerations at the allelic level
Considering the complexity of the procedure and the threats to the genetic integrity of the accessions, the changes that were observed can be considered small. There was no apparent loss of diversity, and the genetic differentiation was only in two out of six cases significant, even with thirty plants measured before and thirty after regeneration (Table 3). These differences between the accessions before and after regeneration are thus mainly due to sampling effects. This is supported by the dendrograms showing the individual plants in the regenerations (not shown) in which, in all six cases, plants did not cluster according to generation.
The changes over regenerations were hardly significant at the population level, but at the allelic level there were some very clear frequency changes (Fig. 2). These changes were similar to the differences between similar accessions within a group such as the differences between the two most similar accessions found in the study CGN07039 and CGN14091 (Fig. 2). Three causes of these differences should be considered, i.e., methodological errors, random processes and effects of selection. Concerning the methodological errors, the reproducibility was shown to be very high. Occasional errors due to contamination of plants with fungi or other organisms or occasional irreproducibility of PCR product formation in AFLP analysis are possible, but given the consistency of the occurrence of significantly different bands not very probable. Concerning the random processes, both genetic sampling effects during the regeneration and statistical sampling effects during the analysis should be considered. The observed differences were too extreme, however, to be explained by either of those. Genetic sampling effects result in a continuous distribution of χ2 values, and do not explain the high values that were observed in all regenerations. Statistical sampling effects, due to the high number of χ2 values calculated, could have been corrected with the Bonferroni correction but this would not change the results as was described earlier. What remains is the third possible cause: the differences might be the result of close genetic linkage of these loci to QTL’s or qualitative traits that were selected during regeneration. However, the fact that the significant differences are caused by different bands in different regenerations does not support this hypothesis. In the regeneration of CGN11160, an additional explanation is possible, which is related to the relatively high level of homogeneity in this accession (cf. Table 3). Inbreeding resulting in reduced heterogeneity within the accession, might have reached a level at which problems in subsequent multiplication occur, i.e. matings leading to successful seed set are no longer random due to incompatibility factors. As a consequence, significant shifts in band frequencies can occur in the next generation. In Fig. 2, this is not only observed in the occurrence of band frequency shifts with relatively high χ2 values, but also in a relative lack of band frequency shifts with a χ2 of 1 or lower compared to the distribution of χ2 values in the other regenerations presented in Fig. 2. This pattern is also seen in the CGN07039-14091 comparison (Fig. 2), where the same explanation could apply because of a similar level of genetic diversity in these accessions (Hs of 0.086 and 0.089, respectively).
Changes over regenerations versus differences between accessions
When the genetic diversity between accessions in the collection is studied, some differences between individual accessions are surprisingly small (Table 2, Fig. 1). The accessions within the duplication groups ‘Gouden Akker’ and ‘Herfstdeen’ (latter not shown) are so closely related that one regeneration would result in comparable genetic differences, also if the amount of diversity within the accessions is taken into consideration. Some other groups, such as the ‘Langedijker’ group, which is shown as reference in Fig. 1, show considerable larger differences, even comparable to the differences in the ‘Dutch genepool’ or the ‘world genepool’ groups. These groups should therefore not be considered as duplication groups.
The small differences between accessions as related to the differences over regenerations can be interpreted as an indication that the regeneration protocols should be improved. This improvement should be aimed at reduction of selection, since the important changes that were observed were not caused by drift, but only by shift. Selection can be reduced, for example, by harvesting equal amounts of seeds per plant, or by controlled pollination. Such measures, however, will generally be quite costly, and might not be feasible economically.
The small differences between accessions as related to the differences over regenerations can also be interpreted as an indication that these similar accessions should be combined in a new bulk accession. Some of the accessions in the CGN Brassica collection are already the result of such combination of very similar samples prior to inclusion in the collection (Hintum et al. 1996). Based on the advice of crop experts, who studied the passport data and who looked at the material in the field different selections from common landraces were merged in one or more groups. This resulted in a considerable reduction of the number of (candidate) accessions. This effort of merging material was supported by a study using isozymes, which concluded that the groups might even have been larger (Hintum et al. 1996). This conclusion is supported by the current study: material of groups such as the ‘Gouden Akker’ could have been further merged based on the observed diversity. Phippen et al. (1997) also concluded, in their case on the basis of a RAPD study on the ‘Golden Acre’ group (same as ‘Gouden Akker’), that the number of accessions could be reduced without significant loss of diversity. It should, however, be taken into account that neutral markers such as AFLPs, as used in the current study and RAPDs, as used by Phippen et al. (1997), show mainly neutral diversity whereas germplasm users are interested in functional diversity. The decisions of not combining certain accessions have always been based on the functional diversity they possessed. Based on expert advice it was often decided to keep the distinct morphological or phenological types separate: the early, the flat or the white. The question how difficult it would be to reconstruct such types via simple mass selection in the combined accession is relevant in this respect. If gene banks would be able to reduce costs without losing genetic diversity by combining genetically highly similar accessions of cross-pollinating crops, distinct phenotypes of morphological or phenological traits might be lost. One could recover these phenotypes by one or two cycles of mass selection, but this would have to be done by the user of the material. This implies that quality, in the form of phenotypes, would be sacrificed to save money. However, this money could be invested with a higher return in other quality aspects of gene bank management.
Conclusion
The genetic changes over normal gene bank regenerations, as measured by AFLPs, are of a comparable magnitude as the differences between some of the more similar accessions. These changes and differences are mainly due to some highly significant differences in allele frequencies, whereas the majority of alleles occur in similar frequencies. This suggests that selection is taking place. This implies that either regeneration protocols should be improved or the composition of the collection should be changed either by merging, removing or replacing gene bank accession.
References
Bland JM, Altman DG (1995) Multiple significance tests: the Bonferroni method. Br Med J 310:170
Bonferroni CE (1936) Teoria statistica delle classi e calcolo delle probabilita. Pubblicazioni del R Istituto Superiore di Scienze Economiche e Commerciali di Firenze 8:3–62
Borner A, Chebotar S, Korzun V (2000) Molecular characterization of the genetic integrity of wheat (Triticum aestivum L.) germplasm after long-term maintenance. Theor Appl Genet 100:494–497
Boukema IW, Hintum TJL van (1998) The European Brassica database. Proceedings of an international symposium on Brassicas. Acta Hort 459:249–254
Chebotar S, Röder MS, Korzum V, Saal B, Weber WE, Borner A (2003) Molecular studies on genetic integrity of open-pollinating species rye (Secale cereale L.) after long-term gene bank maintenance. Theor Appl Genet 107:1469–1476
Crow JF (1986) Basic concepts in population, quantitative and evolutionary genetics. W.H. Freeman, New York, p 273
Diaz O, Gustafsson M, Astley D (1997) Effect of regeneration procedures on genetic diversity in Brassica napus and B. rapa as estimated by isozyme analysis. Genet Resour Crop Evol 44:523–532
FAO (1996) FAO State of the World’s plant genetic resources for food and agriculture. Rome
Falconer DS (1981) Introduction to quantitative genetics 2nd edn. Longman, London
Fulton TM, Chunwongse J, Tanksley SD (1995) Microprep protocol for extraction of DNA from tomato and other herbaceous plants. Plant Mol Biol Rep 13:207–209
Hintum TJL van, Boukema IW, Visser DL (1996) Reduction of duplication in a Brassica oleracea germplasm collection. Genet Res Crop Evol 43:343–349
Hintum TJL van, Brown AHD, Spillane C, Hodgkin T (2000) Core collections of plant genetic resources. IPGRI Technical Bulletin No. 3. International Plant Genetic Resources Institute, Rome, p 48
Hintum TJL van, Sackville Hamilton NR, Engels JMM, Treuren R van (2002) Accession management strategies: splitting and lumping. In: Managing plant genetic resources. CAB International 2002, pp 113–120
McGregor CE, Treuren R van, Hoekstra R, Hintum TJL van (2002) Analysis of the wild potato germplasm of the series Acaulia with AFLPs: implications for ex situ conservation. Theor Appl Genet 104:146–156
Nei M (1975) Molecular population genetics and evolution. Elsevier, New York, p 356
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Parzies HK, Spoor W, Ennos RA (2000) Genetic diversity of barley landrace accessions (Hordeum vulgare ssp. vulgare) conserved for different lengths of time in ex situ gene banks. Heredity 84:476–486
Perneger TV (1998) What is wrong with Bonferroni adjustments. Br Med J 136:1236–1238
Phippen WB, Kresovich S, Candelas FG, McFerson JR (1997) Molecular characterization can quantify and partition variation among gene bank holdings: a case study with phenotypically similar accessions of Brassica oleracea var. capitata L. (cabbage) ‘Golden Acre’. Theor Appl Genet 94:227–234
Reedy ME, Knapp AD, Lamkey KR (1995) Isozyme allelic frequency changes following maize (Zea mays L.) germplasm regeneration. Maydica 40:269–273
Rogers JS (1972) Measures of genetic similarity and genetic distance. In: Studies in Genetics VII. University of Texas Publication 7213, Austin
Sackville Hamilton NR, Engels JMM, Hintum TJL van, Koo B, Smale M (2002) Accession management. Combining or splitting accessions as a tool to improve germplasm management efficiency. IPGRI Technical Bulletin No. 5. International Plant Genetic Resources Institute, Rome
Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414
Wright S (1978) Evolution and the genetics of populations, vol. 4: Variability within and among natural populations. University of Chicago Press, Chicago
Acknowledgments
The authors would like to thank Ietje Boukema and Liesbeth de Groot for their collaboration in selecting and supplying the material analysed in this experiment, and Rene Smulders and Bert Visser for their detailed comments that greatly improved the quality of the manuscript. The authors are also grateful for the valuable suggestions for improvement given by the reviewers of the paper.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by H. H. Geiger.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License ( https://creativecommons.org/licenses/by-nc/2.0 ), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
van Hintum, T.J.L., van de Wiel, C.C.M., Visser, D.L. et al. The distribution of genetic diversity in a Brassica oleracea gene bank collection related to the effects on diversity of regeneration, as measured with AFLPs. Theor Appl Genet 114, 777–786 (2007). https://doi.org/10.1007/s00122-006-0456-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-006-0456-2