Abstract
Recently discovered microorganisms affiliated to the bacterial phylum NC10, named “Candidatus Methylomirabilis oxyfera”, perform nitrite-dependent anaerobic methane oxidation. These microorganisms could be important players in a novel way of anaerobic wastewater treatment where ammonium and residual dissolved methane might be removed at the expense of nitrate or nitrite. To find suitable inocula for reactor startup, ten selected wastewater treatment plants (WWTPs) located in The Netherlands were screened for the endogenous presence of M. oxyfera using molecular diagnostic methods. We could identify NC10 bacteria with 98% similarity to M. oxyfera in nine out of ten WWTPs tested. Sludge from one selected WWTP was used to start a new enrichment culture of NC10 bacteria. This enrichment was monitored using specific pmoA primers and M. oxyfera cells were visualized with fluorescence oligonucleotide probes. After 112 days, the enrichment consumed up to 0.4 mM NO −2 per day. The results of this study show that appropriate sources of biomass, enrichment strategies, and diagnostic tools existed to start and monitor pilot scale tests for the implementation of nitrite-dependent methane oxidation in wastewater treatment at ambient temperature.
Similar content being viewed by others
Introduction
Anaerobic nitrite-dependent methane oxidation is a recently discovered process performed by bacteria with doubling times of approximately 1–2 weeks (Raghoebarsing et al. 2006; Ettwig et al. 2008). The dominant bacteria present in the anaerobic enrichment cultures were members of the NC10 phylum (Ettwig et al. 2009; Hu et al. 2009). The genome of this dominant bacterium, named “Candidatus Methylomirabilis oxyfera”, could be assembled from metagenomic data resulting in a 2.7-Mb circular single chromosome which contained genes of both anaerobic and, surprisingly, aerobic metabolic pathways (Ettwig et al. 2010). The genome harbored the complete aerobic pathway to oxidize methane, including the pmoCAB operon encoding the particulate methane monooxygenase (pMMO) complex for which recently PCR primers were developed (Luesken et al. 2011). Conversely, the denitrification pathway was not complete. Genes nosDFYZ encoding nitrous oxide reductase were missing from the genome and nosL appeared not to be expressed, indicating that another denitrifying pathway had to be operative (Wu et al. 2011). Dedicated stable isotope studies showed that this organism could make its own molecular oxygen from nitrite via nitric oxide (Ettwig et al. 2010). The produced oxygen was mainly used to oxidize methane in an anaerobic environment according to the expected stoichiometry: \( 3\;{\text{C}}{{\text{H}}_4} + 8\;{\text{N}}{{\text{O}}_2}^{ - } + 8\;{{\text{H}}^{ + }} \to 3\;{\text{C}}{{\text{O}}_2} + 4\;{{\text{N}}_2} + 10\;{{\text{H}}_2}{\text{O}} \) (Eq. 1) (Wu et al. 2011).
Anaerobic wastewater treatment compared to conventional aerobic processes has advantages like a reduced production of sludge, a smaller footprint, and the production of biogas (methane) which can be used as an energy source (van Haandel and Lettinga 1994; Lema and Omil 2001; Aiyuk et al. 2006). One of the most established anaerobic techniques is the upflow anaerobic sludge blanket (UASB) first described by Lettinga et al. (1980). In the absence of oxygen, the microbial community, present in the UASB reactor, degrades organic matter eventually into the main products ammonium and methane (Toerien and Hattingh 1969). The produced ammonium should be removed in accordance with the stringent rules for nitrogen compounds in wastewater effluent (http://ec.europa.eu/environment/water/water-urbanwaste/index_en.html). Methane contributes to the greenhouse effect when released to the environment and should therefore be removed or used in an energy-efficient way (Cakir and Stenstrom 2005; Bogner et al. 2008). M. oxyfera-type bacteria could use methane to drive denitrification, circumventing the purchase of electron donors like methanol for nitrogen removal. To obtain sufficient oxidized nitrogen for methane oxidation by M. oxyfera-type bacteria, partial nitrification of ammonium could be used (van Dongen et al. 2001). This makes M. oxyfera-type bacteria important candidates for a novel way of sustainable anaerobic wastewater treatment.
To date, enrichment cultures of M. oxyfera have been obtained from two different freshwater systems in The Netherlands (Raghoebarsing et al. 2006; Ettwig et al. 2009). In addition, enrichments of NC10 bacteria were obtained from a mixed sample of freshwater sediment, anaerobic sludge, and return activated sludge (Hu et al. 2009). In the present study, we detected M. oxyfera-type bacteria in nine out of ten screened wastewater treatment plants (WWTPs) located in The Netherlands using 16S rRNA gene analysis. One of these WWTPs was selected and biomass was used to start an enrichment culture of M. oxyfera. At the end of the experimental period, this enrichment was capable of nitrite-dependent methane oxidation with conversion rates of 0.3 nmol CH4 min−1 mg protein−1 and 0.9 nmol NO −2 min−1 mg protein−1 at ambient temperature.
Materials and methods
Sampling sites
All the WWTP sludge samples used for screening came from plants located in The Netherlands (Table 1). The WWTPs were selected based on a sludge retention time ≥8 days and BOD/N ≤4.7. The WWTP in Lieshout treats industrial water using UASB reactors followed by a Carrousel. The other nine WWTPs in our selection process wastewater originating from both domestic and industrial sources. These treatment plants make use of pre-denitrification in combination with an activated sludge system. Besides this system, reject water of a digester is processed by the WWTP in Kralingseveer and the WWTP in Lichtenvoorde receives effluent of a process where anaerobically treated wastewater of a tannery is combined with partial nitrification and anaerobic ammonium oxidation (anammox). The WWTPs Haarlo, Varsseveld, and Heerenveen apply chemical phosphorus removal. Samples from the activated sludge were taken for molecular analysis.
The industrial water treatment plant in Lieshout (Bavaria B.V., Lieshout, The Netherlands) uses three separate UASB reactors. Subsequently, the UASB effluent is treated by a Carrousel with two aeration points. The sludge samples for molecular screening and inoculation of a bioreactor were taken at the end of the Carrousel after the second aeration point. Sludge (2 l) was mixed with ambient water to inoculate the bioreactor.
Molecular analysis
DNA from the Lieshout WWTP and the enrichment was extracted and purified according to Ettwig et al. (2009). DNA from the other nine WWTP samples was extracted using the PowerSoil® DNA isolation kit (MO BIO, Carlsbad, CA, USA). Finally, DNA was dissolved in diethylpyrocarbonate (DEPC) treated water (Invitrogen UK) and the quality was checked with gel agarose electrophoresis.
To analyze biodiversity of NC10 phylum bacteria, the isolated DNA was used as a template for a 16S rRNA targeted PCR. The 16S rRNA primers used in the nested PCR approach were NC10 specific forward primer 202F (Ettwig et al. 2009) and general bacterial reverse primer 1545R (Juretschko et al. 1998). Thermal cycling was performed with an initial melting step for 4 min at 94 °C, followed by 35 cycles of denaturation at 94 °C for 1 min, annealing at 57 °C for 1 min, and elongation at 72 °C for 2.5 min. Finally, an elongation step at 72 °C for 10 min was performed. The PCR product obtained was used as template for a nested PCR using the NC10 specific primers qP1F and qP2R, originally developed for qPCR (Ettwig et al. 2009). In the nested PCR, the thermal cycling was performed as described above, with an annealing temperature of 65 °C for 1 min. For the clone library of the 332-day Lieshout enrichment, we used an annealing temperature gradient (50–60 °C).
To identify the M. oxyfera-type bacteria on a functional level, four different primer combinations targeting the pmoA gene were used. Four different DNA samples originating from the inoculum (inoc) and enrichment (enr) of the Lieshout treatment plant (inoc Lieshout, enr Lieshout, enr 332 day Lieshout, and WWTP Lieshout) served as a template for PCR and subsequently pmoA gene libraries were constructed. First, novel forward primer A189_b (Luesken et al. 2011) with general reverse primer 682R (Holmes et al. 1995) were used in a direct PCR. Secondly, the forward primer A189_b and reverse primer cmo682 (Luesken et al. 2011) were used in a nested PCR approach with M. oxyfera specific primers cmo182 and cmo568 (Luesken et al. 2011). The third primer combination was forward primer A189_b combined with reverse primer cmo682, and in the fourth combination primers cmo182 and cmo568 were used. The third and fourth primer combinations were used in a direct PCR on DNA samples of the enrichment obtained from Lieshout sludge. For all the four primer combinations used, thermal cycling was performed according to Luesken et al. (2011). All PCR reactions were performed with the Quanta BioScience Inc., PerfeCTa® SYBR® Green FastMix® 76 (Gaithersburg, MD, USA).
The PCR products were cloned with the pGEM®-T Easy cloning kit (Promega USA) and XL1 Blue Escherichia coli competent cells. A maximum of about 20 clones per library were selected. Subsequently, the plasmids were isolated using the GeneJet miniprep kit (Fermentas, Lithuania). The Diagnostics Center of Nijmegen University Medical Center performed sequencing using M13 forward and reverse primers. The quality of the sequences was checked using the Chromas LITE (version 2.01) program. To obtain related sequences (>93% similarity) from GenBank (http://www.ncbi.nlm.nih.gov/GenBank/), a BLAST search was performed. Representative 16S rRNA and pmoA sequences from each location were submitted to GenBank (accession numbers JF706170–JF706214). The sequences were aligned using MEGA4 software (Tamura et al. 2007), and manually checked and trimmed. Phylogenetic trees were calculated in MEGA with the neighbor-joining method and with the pairwise deletion option for gaps. The tree topology was tested by bootstrap analysis (1,000 replicates).
Enrichment and cultivation
Bioreactors of 15 l and 3 l (Applikon, Schiedam, The Netherlands) were used for cultivation. For efficient biomass retention, the bioreactors were operated in a sequencing-batch mode (Strous et al. 1998). The settling cycle consisted of 23.2 h of constant medium supply, 20 min of settling, and 30 min of pumping out the excess liquid. Poorly settling biomass was collected using an external settler (2.3 l). During medium supply (flow rate 0.3–1 l day−1), the reactor was stirred at 100 rpm and sparged with CH4–CO2 (95:5 vol/vol; purity >99.995%; flow rate 10 ml min−1; Air Liquide, France). The bioreactors were equipped with a level controller.
The medium, in both reactor systems, was continuously sparged with Ar–CO2 (95:5, vol/vol) to maintain anoxic conditions and contained the following components (per liter): KHCO3, 0.5 to 1 g; KH2PO4, 0.05 g; CaCl2·2H2O, 0.3 g; MgSO4·7H2O, 0.2 g; NaNO3, 0.035 to 0.213 g (0.5 to 3 mM); NaNO2, 0.0069 to 0.41 g (0.1 to 6 mM); an acidic trace element solution, 0.5 ml; and an alkaline trace element solution, 0.2 ml. The acidic (100 mM HCl) trace element solution contained (per liter) 2.085 g FeSO4·7H2O, 0.068 g ZnSO4·7H2O, 0.12 g CoCl2·6H2O, 0.5 g MnCl2·4H2O, 0.32 g CuSO4, 0.095 g NiCl2·6H2O, and 0.014 g H3BO3. The alkaline (10 mM NaOH) trace element solution contained (per liter) 0.067 g SeO2, 0.050 g Na2WO4·2H2O, and 0.242 g Na2MoO4. All medium components were sterilized, either by 0.2 μm filtration (acidic trace element solution) or by autoclaving.
Both bioreactors (15 l and 3 l) were equipped with a Clark-type oxygen electrode and pH electrode (continuously monitored) and kept at ambient temperature (20–23 °C). The pH of the cultures liquid varied between 6.8 and 7.3. The bioreactors were wrapped in black foil and black Viton and Norprene tubing with low oxygen permeability was used (Cole Parmer, USA).
When the nitrite concentration was <0.1 or >1 mM, the medium flow was adjusted (influent varied between 0.3 and 1 l day−1). In addition, NaNO2 and KHCO3 concentrations in the medium varied depending on the denitrifying activity of the culture. Both bioreactors were operated aseptically.
Activity measurements
The external settler (2.3 l Schott bottle), coupled to the 15-l bioreactor, was removed from the effluent tubing and operated as batch incubation. To obtain a headspace of ~0.3 l, Ar/CO2 was pumped into the settler and the excess fluid was discarded. Two liters of liquid culture remained in the settler and CH4 was added until a final concentration in the headspace of 4%. On t = 0 h, 10 mg l−1 NO −2 and 100 mg l−1 NO −3 were present in the settler. Samples were taken on a regular basis until 68 h.
To determine the activity in the 3-l bioreactor, batch incubations were performed at t = 308 and 315 days. The methane flow was stopped and the headspace (400 ml) of the bioreactor was flushed with Ar/CO2 (10 ml min−1) for 30 min yielding a CH4 concentration in the headspace of 5.3% and 4.8%, respectively. For the incubation at t = 308 days, the medium flow was stopped and nitrite was added to a final concentration of 1.3 mM in the bioreactor. Measurements were started after a stabilization period (overnight). At t = 315 days, a second activity test was performed with an initial nitrite concentration of 0.2 mM. In both incubations, methane, nitrite, and nitrate samples were taken (until t = 10 h) and analyzed as described below.
Fluorescence in situ hybridization
Biomass samples (2 ml) were taken from the enrichment culture and centrifuged. The pellet was washed in phosphate-buffered saline (PBS; 10 mM Na2HPO4/NaH2PO4 pH 7.5 and 130 mM NaCl) and fixed for 2 h on ice in 3% (w/v) paraformaldehyde in PBS. After incubation, the samples were washed with PBS and resuspended in ethanol and PBS (1:1). The fixed sample was stored at −18 °C.
Fixed biomass (10 μl) was spotted on Teflon-coated microscopic slides and dehydrated for 3 min in subsequently 50%, 80%, and 96% ethanol. The probes were hybridized for 1.5 h at 46 °C in hybridization buffer (900 mM NaCl, 2 mM Tris/HCl pH 8.0, 0.2‰ sodium dodecyl sulfate) and 50% formamide. The following oligonucleotide probes were used: S-*-DBACT-0193-a-A-18 (DBACT193) and S-*-DBACT-1027-a-A-18 (DBACT1027), specific for bacteria affiliated with the NC10 phylum (Raghoebarsing et al. 2006); and S-D-Bact-0338-a-A-18 (EUB338), specific for most Bacteria (Amann et al. 1990). The slides were examined using a Zeiss Axioplan II epifluorescence microscope with digital video camera and image analysis software (Axiovision, Zeiss, Germany).
Analytical methods
For routine nitrite analysis, Merckoquant test strips (0 to 80 mg l−1 nitrite; Merck, Germany) were used. Nitrite and nitrate samples (1 ml) from the batch experiments were measured colorimetrically (Kartal et al. 2006). The total protein content was determined by the BCA (bicinchoninic acid assay; Pierce, USA) according to the manufacturer’s protocol. Bovine serum albumin (Thermo Scientific, USA) was used as a standard. The methane concentration was determined by gas chromatography (Ettwig et al. 2008).
Results
Screening for M. oxyfera in WWTPs
To find suitable inocula to start pilot scale tests for application of nitrite-dependent anaerobic methane oxidation, ten WWTPs in The Netherlands were screened with molecular tools (Table 1). All WWTPs treat wastewater originating from both domestic and industrial sources, except the WWTP in Lieshout. This plant treats only industrial (brewery) water at moderate temperatures. Relatively long sludge retention times and low BOD/N ratios were used as criteria to select the WWTPs for screening. These criteria in combination with alternating oxic–anoxic conditions present in all WWTPs might reflect the niche of M. oxyfera bacteria where both methane and oxidized nitrogen compounds are present at the same time. A molecular survey was conducted to screen for M. oxyfera-type bacteria after DNA extraction from the sludge samples of these WWTPs. To detect NC10 bacteria, a direct PCR with NC10 specific primer 202F and general primer 1545R targeting the 16S rRNA gene was performed on one of the WWTP sludge samples (Lieshout). This resulted in a PCR product of the right size, but this product was hardly visible with gel electrophoresis (data not shown). Therefore, a nested PCR approach was performed on all WWTP sludge samples. In nine out of ten WWTPs tested, M. oxyfera-type bacteria could be detected (Fig. 1a). M. oxyfera-type bacteria were not detected in the WWTP in Varsseveld. Recently, PCR primer sets for detection of M. oxyfera pmoA genes were published (Luesken et al. 2011). These sets were used in the later stage of this study (see below) complementary to the 16S rRNA approach.
a Phylogenetic tree of 16S rRNA gene sequences of the NC10 phylum, including sequences of nine different WWTPs located in The Netherlands (bold). The phylogenetic tree was constructed using neighbor-joining and bootstrap analysis of 1,000 replicates. All sequences found are represented in group a of the NC10 phylum (group a, b, c, and d are described in Ettwig et al. 2009). b Phylogenetic tree of pmoA sequences including clones from inoc Lieshout, enr Lieshout, enr 332 day Lieshout, and WWTP Lieshout. The phylogenetic tree was constructed using neighbor-joining and bootstrap analysis of 1,000 replicates. The clones indicated with “a” were obtained with the primers A189_b and 682R, clones indicated with “b” were obtained with a nested approach using primers A189_b and cmo682 as a template for primers cmo182 and cmo568. Clones obtained with a direct PCR using the primers A189_b and cmo682 are indicated with “c”. Clones obtained with primers cmo182 and cmo568 are indicated with “d”
Enrichment of M. oxyfera-type bacteria from wastewater sludge
The industrial wastewater treatment plant in Lieshout was selected to start an enrichment culture. Important treatment processes of this plant are three UASB reactors in parallel followed by a Carrousel for nitrification and denitrification. In the UASB reactor, organic compounds are degraded and the produced methane is collected and used as an energy source by the brewery, but dissolved methane is difficult to recover and remains in the liquid phase. The effluent from the UASB reactors contains reduced nitrogen compounds (0.046 ± 0.014 g l−1 N, of which 0.007 ± 0.007 g l−1 is NH +4 ). This effluent is treated in the Carrousel that makes use of two aeration points, resulting in oxic and anoxic zones where nitrification and denitrification can take place (hydraulic retention time is 4 days). Compared to the other screened WWTPs, the Carrousel in this plant has a prolonged sludge retention time (50 days). The presence of both dissolved methane and oxidized nitrogen compounds (nitrate and nitrite), and the prolonged sludge retention time might favor M. oxyfera bacteria. Samples to start an enrichment culture were taken after the second aeration point in the Carrousel.
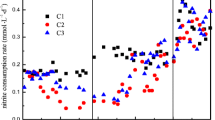
A 15-l sequencing batch reactor was inoculated with biomass from the WWTP Lieshout. During the initial trial period, we noticed that the denitrifying activity measured inside the bioreactor gradually decreased. Furthermore, biomass that was not efficiently retained in the reactor accumulated in the external settler. The biomass collected in the external settler (0.7 l liquid culture containing 1.5 g of protein) did show denitrifying activity: NO −2 and NO −3 were converted at rates of 2.1 and 9.2 nmol h−1 mg protein−1, respectively. Besides the denitrification, anaerobic methane oxidizing activity (1.7 nmol CH4 h−1 mg protein−1) could be measured. In addition to these activity experiments, the biomass in the external settler was analyzed for the presence of NC10 bacteria using a nested PCR approach. M. oxyfera-type bacteria were detected in the settler biomass and were all clustering in group a of the NC10 phylum (Ettwig et al. 2009). The sequences obtained from the external settler were closely related to the 14 sequences found in the inoculum originating from the WWTP in Lieshout (Fig. 1a). Based on these results, the biomass from the external settler was used to inoculate a new 3-l bioreactor (SBR) that was operated at ambient temperature under strict anoxic conditions and a stringent biomass retention regime. At the startup of the 3-l bioreactor, the initial nitrite concentration (1 mM) was diluted to 0.01 mM by pumping fresh medium into the reactor (day 0–50). The activity of the culture till 77 days was low but thereafter the culture showed increasing denitrifying activity up to 0.4 mM of NO −2 day−1 on t = 112 days (Fig. 2a). There were a number of technical problems (power failures, interruption of the methane supply) from days 112 to 175 causing fluctuating nitrite and methane consumption activities. However, from day 175, there was an ongoing increasing nitrite reducing activity up to 1.1 mM day−1 (Fig. 2a).
Enrichment and activity of a culture performing nitrite-dependent anaerobic methane oxidation. a Nitrite reduced by the whole enrichment culture present in a 3-l bioreactor (SBR). After an increase in activity (days 77–112) several technical problem occurred but from day 175 onwards activity gradually increased to 1.1 mM of NO −2 consumed per day. b Methane and nitrite conversion of the whole culture at 308 days. Methane was converted with 0.3 nmol CH4 mg protein−1 min−1 (closed diamonds) and nitrite with 0.9 nmol NO −2 mg protein−1 min−1 (open squares)
Nitrite-dependent anaerobic methane oxidation
Activity measurements were performed with the enrichment culture on t = 308 and 315 days. The anaerobic conversion of nitrite and methane was measured in the 3-l bioreactor. At that time, the protein concentration in the enrichment culture was 0.4 g l−1. Nitrite was added to the enrichment culture prior to the experiment with a final concentration of 1.3 mM. The measured anaerobic methane oxidizing activity was 0.3 nmol CH4 min−1 mg protein−1. Nitrite was converted with 0.9 nmol NO −2 min−1 mg protein−1 (Fig. 2b). In this study, the measured stoichiometry for methane to nitrite was 3 CH4:9.8 NO −2 , this value was close to theoretical value 3 CH4:8 NO −2 (Raghoebarsing et al. 2006; Ettwig et al. 2009). After 315 days, the conversion rates for methane and nitrite were 0.5 nmol min−1 mg protein−1 and 1.2 nmol min−1 mg protein−1, respectively (data not shown). The measured stoichiometry was 3 CH4:10.1 NO −2 .
Identification of M. oxyfera-type bacteria
To investigate the biodiversity in the enrichment after 332 days, the culture was analyzed using specific NC10 primers targeting the 16S rRNA gene. The sequences obtained from this enriched biomass were highly similar to the 16S rRNA gene sequences found in the inoculum and were all clustering in group a of the NC10 phylum (Ettwig et al. 2009). During cultivation, the NC10 members of group a were predominantly enriched (Fig. 1a). Similar observations were made in a previous study (Ettwig et al. 2009) and suggested that the NC10 bacteria present in group a performed nitrite-dependent anaerobic methane oxidation. In addition to the phylogenetic analysis after 332 days, samples directly taken from the Lieshout WWTP (1 year after taking the inoculum) were screened. Sequences retrieved from the WWTP in Lieshout (six clones), 1 year after taking the inoculum, were all clustering in group b of the NC10 phylum (Ettwig et al. 2009). There were no sequences found in group a of the NC10 phylum, unlike the results of the original inoculum where 14 sequences clustered in group a (Fig. 1a).
Since the genome of M. oxyfera contained the complete pathway to oxidize methane aerobically, pmoA could serve as a functional marker. The pmoA gene encodes one of the subunits of the pMMO complex that facilitates the aerobic conversion of methane to methanol. Specific pmoA primers were recently developed to monitor and identify NC10 bacteria on a functional level (Luesken et al. 2011). The four different DNA samples (originating from the Lieshout treatment plant) analyzed for 16S rRNA were also used as a template testing four different primer combinations targeting the pmoA gene. First, a newly developed primer A189_b (Luesken et al. 2011) was combined with the commonly used 682R primer (Holmes et al. 1995). All pmoA sequences retrieved in this study using this primer combination (clone libraries indicated with “a”), clustered with either Methylococcus capsulatus (Gammaproteobacteria) or Crenothrix polyspora (Gammaproteobacteria) and not with M. oxyfera (Fig. 1b). Secondly, the primer combination A189_b and cmo682 was used in a nested PCR approach with novel primers cmo182 and cmo568 as described by Luesken et al. (2011) to detect pmoA sequences related to M. oxyfera. All four DNA samples were analyzed using this combination (clone libraries indicated with “b”) and contained sequences clustering with pmoA gene sequences of M. oxyfera (Fig. 1b). In addition to these two experiments, clone libraries for the detection of M. oxyfera in the enrichment were made using a direct specific PCR with either primer combination A189_b and cmo682 (clone libraries indicated with “c”) or cmo182 and cmo568 (indicated with “d”). Although the third combination with primers A189_b and cmo682 resulted in a PCR product with settler DNA as the template, only one pmoA sequence (out of 13) was obtained after cloning and sequencing. The DNA extracted after 332 days of enrichment (Fig. 1b) yielded 12 pmoA sequences (out of 12 clones screened). The fourth and last combination was a direct PCR with pmoA primers cmo182 and cmo568 using DNA from the 332-day enrichment as template. All clones obtained were similar to the pmoA gene sequence of M. oxyfera (Fig. 1b).
Fluorescence in situ hybridization
In order to get an impression of the relative abundance of M. oxyfera cells compared to the other community members, fluorescence in situ hybridization (FISH) was performed using general and specific probes (Fig. 3). No M. oxyfera-type bacteria could be detected in the inoculum (Fig. 3a), probably because the number of cells was below detection level (Amann et al. 1995). This is consistent with the 16S rRNA and pmoA phylogenetic analysis (see above). Although PCR is a more sensitive technique compared to FISH, a nested PCR approach was necessary to detect M. oxyfera-type bacteria in the inoculum. After 64 days of cultivation in the 3-l bioreactor, approximately 2–3% of the microbial community hybridized with specific probes for M. oxyfera (Fig. 3b). After 308 days, the M. oxyfera population was enriched for approximately 60–70% (Fig. 3c). The enriched bacteria appeared to grow mainly in clusters, but some single cells were also visible as observed previously (Ettwig et al. 2009).
Increasing population of M. oxyfera-type bacteria in Lieshout enrichment culture, visualized with FISH. The cells were hybridized with the probes S-*-DBACT-1027-a-A-18 specific for the NC10 phylum (red) and S-D-Bact-0338-a-A-18 that target most, but not all bacteria (dark blue). a There were no M. oxyfera cells detected in the inoculum, using FISH. b After an incubation period of 64 days in a 3-l reactor, M. oxyfera-type bacteria were enriched for ~2–3% (represented in pink, caused by double hybridization of DBACT1027 and EUB338). c Clusters and some detached cells of M. oxyfera-type bacteria are present in the culture after 308 days (represented in pink). M. oxyfera-type bacteria were enriched for ~60–70%. Probe S-*-DBACT-0193-a-A-18 specific for M. oxyfera bacteria showed the same result. Scale bar is 20 μm
Discussion
Ten WWTPs were screened with molecular tools to find suitable inocula to start pilot scale tests for application of nitrite-dependent anaerobic methane oxidation. In this study, NC10 specific primers targeting the 16S rRNA gene were used to identify M. oxyfera-type bacteria in the selected WWTPs. It appeared to be that a nested PCR approach was necessary to detect NC10 bacteria in the WWTPs sludge samples. Apparently, it was difficult to amplify the 16S rRNA gene from NC10 bacteria using a direct PCR, probably caused by low amounts of DNA from NC10 bacteria in the tested sludges. This is in contrast with previous studies using freshwater sediment samples, in which a direct PCR with 202F and 1492R resulted in strong PCR bands of the right size and subsequently obtained sequences belonged to the NC10 phylum (Ettwig et al. 2009). Alternatively, it could be that the WWTPs screened in this study harbored NC10 bacteria that have more mismatches with the primers used. These primers are based on a limited amount of NC10 sequences presently known, and therefore could miss members of this phylum which have a (slightly) different nucleotide composition at the primer positions. M. oxyfera-type bacteria were not detected in the WWTP in Varsseveld, which may be caused by severe mismatches of the primers or the prevailing conditions in this system. Varsseveld is the only plant in our selection that makes use of a membrane bio reactor (MBR). Before the biological treatment starts in this MBR, the wastewater is filtered thoroughly. Furthermore, to prevent the membranes from clogging, intensive aeration and periodical chemical cleaning are necessary. This might cause an environment not favorable for NC10 bacteria.
Sludge from the treatment plant in Lieshout was selected to enrich a nitrite-dependent methane oxidizing culture. After 308 and 315 days of enrichment in the 3-l bioreactor, experiments to determine the conversion rates of methane and nitrite were performed. The observed stoichiometry for methane to nitrite was 3 CH4:9.8 NO −2 (308 days) and 3 CH4:10.1 NO −2 (315 days), which is comparable to the theoretical value 3 CH4:8 NO −2 (Raghoebarsing et al. 2006; Ettwig et al. 2009). The slight deviation indicated that there was still additional denitrification. This process may have been using other electron donors than methane like organic compounds or ammonium. Similar observations were made in previous studies (Raghoebarsing et al. 2006; Hu et al. 2009). The measured conversion rates for methane to nitrite in this study are relatively low compared to previous experiments (Raghoebarsing et al. 2006; Ettwig et al. 2008, 2009). It is possible that the protein content in the current enrichment was overestimated since organic compounds of activated sludge interfered with the BCA assay (Ras et al. 2008). Organic compounds might be present in the Lieshout enrichment, as a remainder of the UASB process.
To detect M. oxyfera-type bacteria on a functional level, primers targeting the pmoA gene were used on samples originating from the WWTP Lieshout and the enrichment culture. The new pmoA primer A189_b (Luesken et al. 2011) was combined with the widely applied 682R primer (Holmes et al. 1995). Using this combination, pmoA sequences were found that did not cluster with M. oxyfera but clustered with M. capsulatus (Gammaproteobacteria) or C. polyspora (Gammaproteobacteria). This is in accordance with previous results where it was shown that M. oxyfera has some critical mismatches with, especially, the known pmoA reverse primers (Luesken et al. 2011). With a nested PCR approach, sequences clustering with the pmoA sequence present in the genome of M. oxyfera were retrieved in all tested samples. The sequences detected in the inoculum and in the WWTP 1 year after taking the inoculum were similar to each other and to the sequence of the M. oxyfera pmoA gene. This may indicate that there is a small but persistent population of NC10 bacteria present in this WWTP. Two different pmoA primer combinations were used in a direct PCR on samples of the enrichment originating from the WWTP Lieshout. After 332 days of enrichment, pmoA sequences could be retrieved. These results implied that when M. oxyfera-type bacteria were enriched, a direct PCR with specific pmoA primers can be used.
Conclusively, we showed that nine out of ten selected WWTPs harbor small amounts of M. oxyfera bacteria clustering within the NC10 phylum, using molecular tools for screening and detection of these bacteria. In addition, an enrichment culture (60–70%) from wastewater sludge was obtained performing nitrite-dependent methane oxidation at ambient temperature. The M. oxyfera-type bacteria present in this enrichment were identified and monitored using specific pmoA primers. These data suggested that biomass from WWTP systems could be a potential source to start pilot scale reactors with very efficient biomass retention.
References
Aiyuk S, Forrez I, de Lieven K, van Haandel A, Verstraete W (2006) Anaerobic and complementary treatment of domestic sewage in regions with hot climates—a review. Bioresour Technol 97:2225–2241
Amann RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R, Stahl DA (1990) Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol 56:1919–1925
Amann RI, Ludwig W, Schleifer KH (1995) Phylogenetic identification and in-situ detection of individual microbial-cells without cultivation. Microbiol Rev 59:143–169
Bogner J, Pipatti R, Hashimoto S, Diaz C, Mareckova K, Diaz L, Kjeldsen P, Monni S, Faaij A, Gao Q, Zhang T, Ahmed MA, Sutamihardja RT, Gregory R (2008) Mitigation of global greenhouse gas emissions from waste: conclusions and strategies from the Intergovernmental Panel on Climate Change (IPCC) Fourth Assessment Report. Working Group III (Mitigation). Waste Manag Res 26:11–32
Cakir FY, Stenstrom MK (2005) Greenhouse gas production: a comparison between aerobic and anaerobic wastewater treatment technology. Water Res 39:4197–4203
Ettwig KF, Shima S, van de Pas-Schoonen KT, Kahnt J, Medema MH, Op den Camp HJ, Jetten MSM, Strous M (2008) Denitrifying bacteria anaerobically oxidize methane in the absence of Archaea. Environ Microbiol 10:3164–3173
Ettwig KF, van Alen T, van de Pas-Schoonen KT, Jetten MS, Strous M (2009) Enrichment and molecular detection of denitrifying methanotrophic bacteria of the NC10 phylum. Appl Environ Microbiol 75:3656–3662
Ettwig KF, Butler MK, Le Paslier D, Pelletier E, Mangenot S, Kuypers MM, Schreiber F, Dutilh BE, Zedelius J, de Beer D, Gloerich J, Wessels HJ, van Alen T, Luesken F, Wu ML, van de Pas-Schoonen KT, Op den Camp HJ, Janssen-Megens EM, Francoijs KJ, Stunnenberg H, Weissenbach J, Jetten MSM, Strous M (2010) Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 464:543–548
Holmes AJ, Costello A, Lidstrom ME, Murrell JC (1995) Evidence that particulate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiol Lett 132:203–208
Hu S, Zeng RJ, Burow LC, Lant P, Keller J, Yuan Z (2009) Enrichment of denitrifying anaerobic methane oxidizing microorganisms. Envir Microbiol Rep 1:377–384
Juretschko S, Timmermann G, Schmid M, Schleifer KH, Pommerening-Roser A, Koops HP, Wagner M (1998) Combined molecular and conventional analyses of nitrifying bacterium diversity in activated sludge: Nitrosococcus mobilis and Nitrospira-like bacteria as dominant populations. Appl Environ Microbiol 64:3042–3051
Kartal B, Koleva M, Arsov R, van der Star W, Jetten MS, Strous M (2006) Adaptation of a freshwater anammox population to high salinity wastewater. J Biotechnol 126:546–553
Lema JM, Omil F (2001) Anaerobic treatment: a key technology for a sustainable management of wastes in Europe. Water Sci Technol 44:133–140
Lettinga G, van Velsen AFM, Hobma SW, Dezeeuw W, Klapwijk A (1980) Use of the upflow sludge blanket (USB) reactor concept for biological wastewater-treatment, especially for anaerobic treatment. Biotechnol Bioeng 22:699–734
Luesken FA, Zhu B, van Alen TA, Butler MK, Rodriguez Diaz M, Song B, Op den Camp HJM, Jetten MSM, Ettwig KF (2011) pmoA primers for detection of anaerobic methanotrophs. Appl Environ Microbiol 77:3877–3880
Raghoebarsing AA, Pol A, van de Pas-Schoonen KT, Smolders AJ, Ettwig KF, Rijpstra WI, Schouten S, Damste JS, Op den Camp HJ, Jetten MS, Strous M (2006) A microbial consortium couples anaerobic methane oxidation to denitrification. Nature 440:918–921
Ras M, Girbal-Neuhauser E, Paul E, Sperandio M, Lefebvre D (2008) Protein extraction from activated sludge: an analytical approach. Water Res 42:1867–1878
Strous M, Heijnen JJ, Kuenen JG, Jetten MSM (1998) The sequencing batch reactor as a powerful tool for the study of slowly growing anaerobic ammonium-oxidizing microorganisms. Appl Microbiol Biotechnol 50:589–596
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Toerien DF, Hattingh WH (1969) Anaerobic digestion. I. Microbiology of anaerobic digestion. Water Res 3:385–416
van Dongen U, Jetten MSM, van Loosdrecht MCM (2001) The SHARON((R))–Anammox((R)) process for treatment of ammonium rich wastewater. Water Sci Technol 44:153–160
van Haandel AC, Lettinga G (1994) Anaerobic sewage treatment—a practical guide for regions with a hot climate. Wiley, Chichester
Wu ML, Ettwig KF, Jetten MS, Strous M, Keltjens JT, Niftrik L (2011) A new intra-aerobic metabolism in the nitrite-dependent anaerobic methane-oxidizing bacterium Candidatus 'Methylomirabilis oxyfera'. Biochem Soc Trans 39:243–248
Acknowledgments
We would like to thank Manfred van den Heuvel and Rob Smit for their help with sampling at the industrial water treatment plant of Bavaria N.V. in Lieshout and Fons Stams for helpful discussions. F.L., T.v.A., C.K., and T.H. were supported by the Foundation for Applied Research (STW project 07736). M.S. is supported by ERC grant "MASEM" (242635). M.J. was supported by ERC grant no. 2322937.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Table 1
List of primers used for phylogenetic analysis targeting the 16S rRNA and pmoA genes. (PDF 33 kb)
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Luesken, F.A., van Alen, T.A., van der Biezen, E. et al. Diversity and enrichment of nitrite-dependent anaerobic methane oxidizing bacteria from wastewater sludge. Appl Microbiol Biotechnol 92, 845–854 (2011). https://doi.org/10.1007/s00253-011-3361-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-011-3361-9